Structural basis of soluble membrane attack complex packaging for clearance
- PMID: 34667172
- PMCID: PMC8526713
- DOI: 10.1038/s41467-021-26366-w
Structural basis of soluble membrane attack complex packaging for clearance
Abstract
Unregulated complement activation causes inflammatory and immunological pathologies with consequences for human disease. To prevent bystander damage during an immune response, extracellular chaperones (clusterin and vitronectin) capture and clear soluble precursors to the membrane attack complex (sMAC). However, how these chaperones block further polymerization of MAC and prevent the complex from binding target membranes remains unclear. Here, we address that question by combining cryo electron microscopy (cryoEM) and cross-linking mass spectrometry (XL-MS) to solve the structure of sMAC. Together our data reveal how clusterin recognizes and inhibits polymerizing complement proteins by binding a negatively charged surface of sMAC. Furthermore, we show that the pore-forming C9 protein is trapped in an intermediate conformation whereby only one of its two transmembrane β-hairpins has unfurled. This structure provides molecular details for immune pore formation and helps explain a complement control mechanism that has potential implications for how cell clearance pathways mediate immune homeostasis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
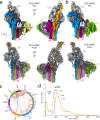


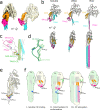
Similar articles
-
Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.J Biol Chem. 2011 May 20;286(20):17585-92. doi: 10.1074/jbc.M111.219766. Epub 2011 Mar 25. J Biol Chem. 2011. PMID: 21454577 Free PMC article.
-
CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.Nat Commun. 2018 Dec 14;9(1):5316. doi: 10.1038/s41467-018-07653-5. Nat Commun. 2018. PMID: 30552328 Free PMC article.
-
Structural basis for membrane attack complex inhibition by CD59.Nat Commun. 2023 Feb 16;14(1):890. doi: 10.1038/s41467-023-36441-z. Nat Commun. 2023. PMID: 36797260 Free PMC article.
-
Soluble Membrane Attack Complex: Biochemistry and Immunobiology.Front Immunol. 2020 Nov 10;11:585108. doi: 10.3389/fimmu.2020.585108. eCollection 2020. Front Immunol. 2020. PMID: 33240274 Free PMC article. Review.
-
How Structures of Complement Complexes Guide Therapeutic Design.Subcell Biochem. 2021;96:273-295. doi: 10.1007/978-3-030-58971-4_7. Subcell Biochem. 2021. PMID: 33252733 Review.
Cited by
-
CFH Haploinsufficiency and Complement Alterations in Early-Onset Macular Degeneration.Invest Ophthalmol Vis Sci. 2024 Apr 1;65(4):43. doi: 10.1167/iovs.65.4.43. Invest Ophthalmol Vis Sci. 2024. PMID: 38683564 Free PMC article.
-
Identification of common and distinct origins of human serum and breastmilk IgA1 by mass spectrometry-based clonal profiling.Cell Mol Immunol. 2023 Jan;20(1):26-37. doi: 10.1038/s41423-022-00954-2. Epub 2022 Nov 29. Cell Mol Immunol. 2023. PMID: 36447030 Free PMC article.
-
Clinical severity classes in COVID-19 pneumonia have distinct immunological profiles, facilitating risk stratification by machine learning.Front Immunol. 2023 Sep 5;14:1192765. doi: 10.3389/fimmu.2023.1192765. eCollection 2023. Front Immunol. 2023. PMID: 37731491 Free PMC article.
-
Complement System Proteins in the Human Aqueous Humor and Their Association with Primary Open-Angle Glaucoma.J Pers Med. 2023 Sep 19;13(9):1400. doi: 10.3390/jpm13091400. J Pers Med. 2023. PMID: 37763167 Free PMC article.
-
The neoepitope of the complement C5b-9 Membrane Attack Complex is formed by proximity of adjacent ancillary regions of C9.Commun Biol. 2023 Jan 13;6(1):42. doi: 10.1038/s42003-023-04431-y. Commun Biol. 2023. PMID: 36639734 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

