Drug repurposing improves disease targeting 11-fold and can be augmented by network module targeting, applied to COVID-19
- PMID: 34667255
- PMCID: PMC8526804
- DOI: 10.1038/s41598-021-99721-y
Drug repurposing improves disease targeting 11-fold and can be augmented by network module targeting, applied to COVID-19
Abstract
This analysis presents a systematic evaluation of the extent of therapeutic opportunities that can be obtained from drug repurposing by connecting drug targets with disease genes. When using FDA-approved indications as a reference level we found that drug repurposing can offer an average of an 11-fold increase in disease coverage, with the maximum number of diseases covered per drug being increased from 134 to 167 after extending the drug targets with their high confidence first neighbors. Additionally, by network analysis to connect drugs to disease modules we found that drugs on average target 4 disease modules, yet the similarity between disease modules targeted by the same drug is generally low and the maximum number of disease modules targeted per drug increases from 158 to 229 when drug targets are neighbor-extended. Moreover, our results highlight that drug repurposing is more dependent on target proteins being shared between diseases than on polypharmacological properties of drugs. We apply our drug repurposing and network module analysis to COVID-19 and show that Fostamatinib is the drug with the highest module coverage.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
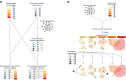
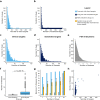
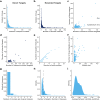
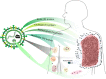
Similar articles
-
Lung disease network reveals impact of comorbidity on SARS-CoV-2 infection and opportunities of drug repurposing.BMC Med Genomics. 2021 Sep 17;14(1):226. doi: 10.1186/s12920-021-01079-7. BMC Med Genomics. 2021. PMID: 34535131 Free PMC article.
-
Repurposing novel therapeutic candidate drugs for coronavirus disease-19 based on protein-protein interaction network analysis.BMC Biotechnol. 2021 Mar 12;21(1):22. doi: 10.1186/s12896-021-00680-z. BMC Biotechnol. 2021. PMID: 33711981 Free PMC article.
-
MNBDR: A Module Network Based Method for Drug Repositioning.Genes (Basel). 2020 Dec 27;12(1):25. doi: 10.3390/genes12010025. Genes (Basel). 2020. PMID: 33375395 Free PMC article.
-
Artificial intelligence in COVID-19 drug repurposing.Lancet Digit Health. 2020 Dec;2(12):e667-e676. doi: 10.1016/S2589-7500(20)30192-8. Epub 2020 Sep 18. Lancet Digit Health. 2020. PMID: 32984792 Free PMC article. Review.
-
Drug Discovery by Drug Repurposing: Combating COVID-19 in the 21st Century.Mini Rev Med Chem. 2021;21(1):3-9. doi: 10.2174/1389557520999200824103803. Mini Rev Med Chem. 2021. PMID: 32838716 Review.
Cited by
-
Network Crosstalk as a Basis for Drug Repurposing.Front Genet. 2022 Mar 8;13:792090. doi: 10.3389/fgene.2022.792090. eCollection 2022. Front Genet. 2022. PMID: 35350247 Free PMC article.
-
Total network controllability analysis discovers explainable drugs for Covid-19 treatment.Res Sq [Preprint]. 2023 Jul 14:rs.3.rs-3147521. doi: 10.21203/rs.3.rs-3147521/v1. Res Sq. 2023. Update in: Biol Direct. 2023 Sep 5;18(1):55. doi: 10.1186/s13062-023-00410-9. PMID: 37503262 Free PMC article. Updated. Preprint.
-
The future of metronomic chemotherapy: experimental and computational approaches of drug repurposing.Pharmacol Rep. 2025 Feb;77(1):1-20. doi: 10.1007/s43440-024-00662-w. Epub 2024 Oct 21. Pharmacol Rep. 2025. PMID: 39432183 Review.
-
Total network controllability analysis discovers explainable drugs for Covid-19 treatment.Biol Direct. 2023 Sep 5;18(1):55. doi: 10.1186/s13062-023-00410-9. Biol Direct. 2023. PMID: 37670359 Free PMC article.
References
-
- Nabirotchkin S, et al. Next-generation drug repurposing using human genetics and network biology. Curr. Opin. Pharmacol. 2019;11:1–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

