The genome-wide rate and spectrum of EMS-induced heritable mutations in the microcrustacean Daphnia: on the prospect of forward genetics
- PMID: 34667306
- PMCID: PMC8626432
- DOI: 10.1038/s41437-021-00478-x
The genome-wide rate and spectrum of EMS-induced heritable mutations in the microcrustacean Daphnia: on the prospect of forward genetics
Abstract
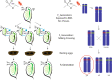
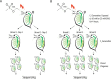
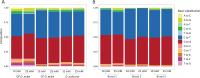
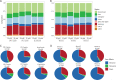
Forward genetic screening using the alkylating mutagen ethyl methanesulfonate (EMS) is an effective method for identifying phenotypic mutants of interest, which can be further genetically dissected to pinpoint the causal genetic mutations. An accurate estimate of the rate of EMS-induced heritable mutations is fundamental for determining the mutant sample size of a screening experiment that aims to saturate all the genes in a genome with mutations. This study examines the genome-wide EMS-induced heritable base-substitutions in three species of the freshwater microcrustacean Daphnia to help guide screening experiments. Our results show that the 10 mM EMS treatment induces base substitutions at an average rate of 1.17 × 10-6/site/generation across the three species, whereas a significantly higher average mutation rate of 1.75 × 10-6 occurs at 25 mM. The mutation spectrum of EMS-induced base substitutions at both concentration is dominated by G:C to A:T transitions. Furthermore, we find that female Daphnia exposed to EMS (F0 individuals) can asexually produce unique mutant offspring (F1) for at least 3 consecutive broods, suggestive of multiple broods as F1 mutants. Lastly, we estimate that about 750 F1s are needed for all genes in the Daphnia genome to be mutated at least once with a 95% probability. We also recommend 4-5 F2s should be collected from each F1 mutant through sibling crossing so that all induced mutations could appear in the homozygous state in the F2 population at 70-80% probability.
© 2021. The Author(s), under exclusive licence to The Genetics Society.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Ethyl methanesulfonate toxicity in Viracept--a comprehensive human risk assessment based on threshold data for genotoxicity.Toxicol Lett. 2009 Nov 12;190(3):317-29. doi: 10.1016/j.toxlet.2009.04.003. Epub 2009 Apr 10. Toxicol Lett. 2009. PMID: 19443141
-
Ethyl methanesulfonate mutant library construction in Gossypium hirsutum L. for allotetraploid functional genomics and germplasm innovation.Plant J. 2020 Jul;103(2):858-868. doi: 10.1111/tpj.14755. Epub 2020 Apr 28. Plant J. 2020. PMID: 32239588
-
Spontaneous and ethyl methanesulfonate-induced mutations controlling viability in Drosophila melanogaster. II. Homozygous effect of polygenic mutations.Genetics. 1977 Nov;87(3):529-45. doi: 10.1093/genetics/87.3.529. Genetics. 1977. PMID: 200526 Free PMC article.
-
A review of the genetic effects of ethyl methanesulfonate.Mutat Res. 1984 Sep-Nov;134(2-3):113-42. doi: 10.1016/0165-1110(84)90007-1. Mutat Res. 1984. PMID: 6390190 Review.
-
Literature review on the genotoxicity, reproductive toxicity, and carcinogenicity of ethyl methanesulfonate.Toxicol Lett. 2009 Nov 12;190(3):254-65. doi: 10.1016/j.toxlet.2009.03.016. Epub 2009 Mar 28. Toxicol Lett. 2009. PMID: 19857796 Review.
Cited by
-
Efficient CRISPR genome editing and integrative genomic analyses reveal the mosaicism of Cas-induced mutations and pleiotropic effects of scarlet gene in an emerging model system.bioRxiv [Preprint]. 2024 Jan 31:2024.01.29.577787. doi: 10.1101/2024.01.29.577787. bioRxiv. 2024. Update in: Heredity (Edinb). 2025 Apr;134(3-4):221-233. doi: 10.1038/s41437-025-00750-4. PMID: 38352317 Free PMC article. Updated. Preprint.
-
The effects of mutations on gene expression and alternative splicing.Proc Biol Sci. 2023 Jul 12;290(2002):20230565. doi: 10.1098/rspb.2023.0565. Epub 2023 Jul 5. Proc Biol Sci. 2023. PMID: 37403507 Free PMC article.
-
Resurrection genomics provides molecular and phenotypic evidence of rapid adaptation to salinization in a keystone aquatic species.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2217276120. doi: 10.1073/pnas.2217276120. Epub 2023 Feb 2. Proc Natl Acad Sci U S A. 2023. PMID: 36730191 Free PMC article.
-
The mosaicism of Cas-induced mutations and pleiotropic effects of scarlet gene in an emerging model system.Heredity (Edinb). 2025 Apr;134(3-4):221-233. doi: 10.1038/s41437-025-00750-4. Epub 2025 Feb 20. Heredity (Edinb). 2025. PMID: 39979422
-
The use of droplet-based microfluidic technologies for accelerated selection of Yarrowia lipolytica and Phaffia rhodozyma yeast mutants.Biol Methods Protoc. 2024 Jul 10;9(1):bpae049. doi: 10.1093/biomethods/bpae049. eCollection 2024. Biol Methods Protoc. 2024. PMID: 39114747 Free PMC article. Review.
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Altshuler I, Demiri B, Xu S, Constantin A, Yan ND, Cristescu ME. An integrated multi-disciplinary approach for studying multiple stressors in freshwater ecosystems: Daphnia as a model organism. Integr Comp Biol. 2011;51:623–633. - PubMed
-
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

