SOMA: Subject-, object-, and modality-adapted precision atlas approach for automatic anatomy recognition and delineation in medical images
- PMID: 34668207
- PMCID: PMC8678400
- DOI: 10.1002/mp.15308
SOMA: Subject-, object-, and modality-adapted precision atlas approach for automatic anatomy recognition and delineation in medical images
Abstract
Purpose: In the multi-atlas segmentation (MAS) method, a large enough atlas set, which can cover the complete spectrum of the whole population pattern of the target object will benefit the segmentation quality. However, the difficulty in obtaining and generating such a large set of atlases and the computational burden required in the segmentation procedure make this approach impractical. In this paper, we propose a method called SOMA to select subject-, object-, and modality-adapted precision atlases for automatic anatomy recognition in medical images with pathology, following the idea that different regions of the target object in a novel image can be recognized by different atlases with regionally best similarity, so that effective atlases have no need to be globally similar to the target subject and also have no need to be overall similar to the target object.
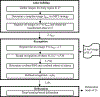
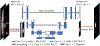
Methods: The SOMA method consists of three main components: atlas building, object recognition, and object delineation. Considering the computational complexity, we utilize an all-to-template strategy to align all images to the same image space belonging to the root image determined by the minimum spanning tree (MST) strategy among a subset of radiologically near-normal images. The object recognition process is composed of two stages: rough recognition and refined recognition. In rough recognition, subimage matching is conducted between the test image and each image of the whole atlas set, and only the atlas corresponding to the best-matched subimage contributes to the recognition map regionally. The frequency of best match for each atlas is recorded by a counter, and the atlases with the highest frequencies are selected as the precision atlases. In refined recognition, only the precision atlases are examined, and the subimage matching is conducted in a nonlocal manner of searching to further increase the accuracy of boundary matching. Delineation is based on a U-net-based deep learning network, where the original gray scale image together with the fuzzy map from refined recognition compose a two-channel input to the network, and the output is a segmentation map of the target object.
Results: Experiments are conducted on computed tomography (CT) images with different qualities in two body regions - head and neck (H&N) and thorax, from 298 subjects with nine objects and 241 subjects with six objects, respectively. Most objects achieve a localization error within two voxels after refined recognition, with marked improvement in localization accuracy from rough to refined recognition of 0.6-3 mm in H&N and 0.8-4.9 mm in thorax, and also in delineation accuracy (Dice coefficient) from refined recognition to delineation of 0.01-0.11 in H&N and 0.01-0.18 in thorax.
Conclusions: The SOMA method shows high accuracy and robustness in anatomy recognition and delineation. The improvements from rough to refined recognition and further to delineation, as well as immunity of recognition accuracy to varying image and object qualities, demonstrate the core principles of SOMA where segmentation accuracy increases with precision atlases and gradually refined object matching.
Keywords: anatomy recognition; multi-atlas segmentation; precision atlas selection.
© 2021 American Association of Physicists in Medicine.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Figures




Similar articles
-
Anatomy Recognition in CT Images of Head & Neck Region via Precision Atlases.Proc SPIE Int Soc Opt Eng. 2021;11596:1159633. doi: 10.1117/12.2581234. Epub 2021 Feb 15. Proc SPIE Int Soc Opt Eng. 2021. PMID: 34887608 Free PMC article.
-
Automatic thoracic anatomy segmentation on CT images using hierarchical fuzzy models and registration.Med Phys. 2016 Mar;43(3):1487-500. doi: 10.1118/1.4942486. Med Phys. 2016. PMID: 26936732
-
Body-wide hierarchical fuzzy modeling, recognition, and delineation of anatomy in medical images.Med Image Anal. 2014 Jul;18(5):752-71. doi: 10.1016/j.media.2014.04.003. Epub 2014 Apr 24. Med Image Anal. 2014. PMID: 24835182 Free PMC article.
-
Multi-atlas image registration of clinical data with automated quality assessment using ventricle segmentation.Med Image Anal. 2020 Jul;63:101698. doi: 10.1016/j.media.2020.101698. Epub 2020 Apr 18. Med Image Anal. 2020. PMID: 32339896 Free PMC article. Review.
-
Taxonomy of Acute Stroke: Imaging, Processing, and Treatment.Diagnostics (Basel). 2024 May 19;14(10):1057. doi: 10.3390/diagnostics14101057. Diagnostics (Basel). 2024. PMID: 38786355 Free PMC article. Review.
References
-
- Cootes TF, Taylor CJ, Cooper DH. Active shape models-their training and application. Comput Vis Image Underst. 1995;61(1): 38–59.
-
- Shen T, Li H, Huang X. Active volume models for medical image segmentation. IEEE Trans Med Imaging. 2011;30(3):774–791. - PubMed
-
- Staib LH, Duncan JS. Boundary finding with parametrically deformable models. IEEE Trans Pattern Anal Mach Intell. 1992;14:1061–1075.

