Transcriptomic and rRNA:rDNA Signatures of Environmental versus Enteric Enterococcus faecalis Isolates under Oligotrophic Freshwater Conditions
- PMID: 34668732
- PMCID: PMC8528121
- DOI: 10.1128/Spectrum.00817-21
Transcriptomic and rRNA:rDNA Signatures of Environmental versus Enteric Enterococcus faecalis Isolates under Oligotrophic Freshwater Conditions
Abstract
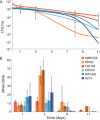
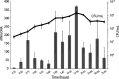
The use of enterococci as a fecal indicator bacterial group for public health risk assessment has been brought into question by recent studies showing that "naturalized" populations of Enterococcus faecalis exist in the extraenteric environment. The extent to which these naturalized E. faecalis organisms can confound water quality monitoring is unclear. To determine if strains isolated from different habitats display different survival strategies and responses, we compared the decay patterns of three E. faecalis isolates from the natural environment (environmental strains) against three human gut isolates (enteric strains) in laboratory mesocosms that simulate an oligotrophic, aerobic freshwater environment. Our results showed similar overall decay rates between enteric and environmental isolates based on viable plate and quantitative PCR (qPCR) counts. However, the enteric isolates exhibited a spike in copy number ratios of 16S rRNA gene transcripts to 16S rRNA gene DNA copies (rRNA:rDNA ratios) between days 1 and 3 of the mesocosm incubations that was not observed in environmental isolates, which could indicate a different stress response. Nevertheless, there was no strong evidence of differential gene expression between environmental and enteric isolates related to habitat adaptation in the accompanying mesocosm metatranscriptomes. Overall, our results provide novel information on how rRNA levels may vary over different growth conditions (e.g., standard lab versus oligotrophic) for this important indicator bacteria. We also observed some evidence for habitat adaptation in E. faecalis; however, this adaptation may not be substantial or consistent enough for integration in water quality monitoring. IMPORTANCE Enterococci are commonly used worldwide to monitor environmental fecal contamination and public health risk for waterborne diseases. However, closely related enterococci strains adapted to living in the extraenteric environment may represent a lower public health risk and confound water quality estimates. We developed an rRNA:rDNA viability assay for E. faecalis (a predominant species within this fecal group) and tested it against both enteric and environmental isolates in freshwater mesocosms to assess whether this approach can serve as a more sensitive water quality monitoring tool. We were unable to reliably distinguish the different isolate types using this assay under the conditions tested; thus, environmental strains should continue to be counted during routine water monitoring. However, this assay could be useful for distinguishing more recent (i.e., higher-risk) fecal pollution because rRNA levels significantly decreased after 1 week in all isolates.
Keywords: bioinformatics; environmental microbiology; fecal organisms; metatranscriptomics; public health; rRNA; water quality.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

