Evolution and Diversity of Bat and Rodent Paramyxoviruses from North America
- PMID: 34668771
- PMCID: PMC8826806
- DOI: 10.1128/JVI.01098-21
Evolution and Diversity of Bat and Rodent Paramyxoviruses from North America
Abstract
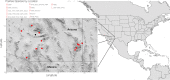
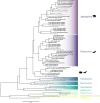
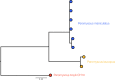
Paramyxoviruses are a diverse group of negative-sense, single-stranded RNA viruses of which several species cause significant mortality and morbidity. In recent years the collection of paramyxovirus sequences detected in wild mammals has substantially grown; however, little is known about paramyxovirus diversity in North American mammals. To better understand natural paramyxovirus diversity, host range, and host specificity, we sought to comprehensively characterize paramyxoviruses across a range of diverse cooccurring wild small mammals in southern Arizona. We used highly degenerate primers to screen fecal and urine samples and obtained a total of 55 paramyxovirus sequences from 12 rodent species and 6 bat species. We also performed Illumina transcriptome sequencing (RNA-seq) and de novo assembly on 14 of the positive samples to recover a total of 5 near-full-length viral genomes. We show there are at least two clades of rodent-borne paramyxoviruses in Arizona, while bat-associated paramyxoviruses formed a putative single clade. Using structural homology modeling of the viral attachment protein, we infer that three of the five novel viruses likely bind sialic acid in a manner similar to other respiroviruses, while the other two viruses from heteromyid rodents likely bind a novel host receptor. We find no evidence for cross-species transmission, even among closely related sympatric host species. Taken together, these data suggest paramyxoviruses are a common viral infection in some bat and rodent species present in North America and illuminate the evolution of these viruses. IMPORTANCE There are a number of viral lineages that are potential zoonotic threats to humans. One of these, paramyxoviruses have jumped into humans multiple times from wild and domestic animals. We conducted one of the largest viral surveys of wild mammals in the United States to better understand paramyxovirus diversity and evolution.
Keywords: Paramyxovirus; codivergence; evolution; mammals.
Figures






References
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

