Susceptibilities of Human ACE2 Genetic Variants in Coronavirus Infection
- PMID: 34668773
- PMCID: PMC8754202
- DOI: 10.1128/JVI.01492-21
Susceptibilities of Human ACE2 Genetic Variants in Coronavirus Infection
Abstract
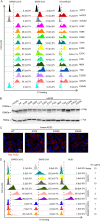
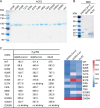
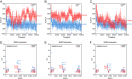
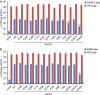
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in more than 235 million cases worldwide and 4.8 million deaths (October 2021), with various incidences and mortalities among regions/ethnicities. The coronaviruses SARS-CoV, SARS-CoV-2, and HCoV-NL63 utilize the angiotensin-converting enzyme 2 (ACE2) as the receptor to enter cells. We hypothesized that the genetic variability in ACE2 may contribute to the variable clinical outcomes of COVID-19. To test this hypothesis, we first conducted an in silico investigation of single-nucleotide polymorphisms (SNPs) in the coding region of ACE2. We then applied an integrated approach of genetics, biochemistry, and virology to explore the capacity of select ACE2 variants to bind coronavirus spike proteins and mediate viral entry. We identified the ACE2 D355N variant that restricts the spike protein-ACE2 interaction and consequently limits infection both in vitro and in vivo. In conclusion, ACE2 polymorphisms could modulate susceptibility to SARS-CoV-2, which may lead to variable disease severity. IMPORTANCE There is considerable variation in disease severity among patients infected with SARS-CoV-2, the virus that causes COVID-19. Human genetic variation can affect disease outcome, and the coronaviruses SARS-CoV, SARS-CoV-2, and HCoV-NL63 utilize human ACE2 as the receptor to enter cells. We found that several missense ACE2 single-nucleotide variants (SNVs) that showed significantly altered binding with the spike proteins of SARS-CoV, SARS-CoV-2, and NL63-HCoV. We identified an ACE2 SNP, D355N, that restricts the spike protein-ACE2 interaction and consequently has the potential to protect individuals against SARS-CoV-2 infection. Our study highlights that ACE2 polymorphisms could impact human susceptibility to SARS-CoV-2, which may contribute to ethnic and geographical differences in SARS-CoV-2 spread and pathogenicity.
Keywords: ACE2; COVID-19; HCoV-NL63; SARS-CoV; SARS-CoV-2; SNP.
Figures


References
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. 10.1056/NEJMoa030781. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

