Sex-Stratified Single-Cell RNA-Seq Analysis Identifies Sex-Specific and Cell Type-Specific Transcriptional Responses in Alzheimer's Disease Across Two Brain Regions
- PMID: 34669146
- PMCID: PMC8786804
- DOI: 10.1007/s12035-021-02591-8
Sex-Stratified Single-Cell RNA-Seq Analysis Identifies Sex-Specific and Cell Type-Specific Transcriptional Responses in Alzheimer's Disease Across Two Brain Regions
Abstract
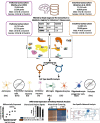
Alzheimer's disease (AD) is a pervasive neurodegenerative disorder that disproportionately affects women. Since neural anatomy and disease pathophysiology differ by sex, investigating sex-specific mechanisms in AD pathophysiology can inform new therapeutic approaches for both sexes. Previous bulk human brain RNA sequencing studies have revealed sex differences in dysregulated molecular pathways related to energy production, neuronal function, and immune response; however, the sex differences in disease mechanisms are yet to be examined comprehensively on a single-cell level. We leveraged nearly 74,000 cells from human prefrontal and entorhinal cortex samples from the first two publicly available single-cell RNA sequencing AD datasets to perform a case versus control sex-stratified differential gene expression analysis and pathway network enrichment in a cell type-specific manner for each brain region. Our examination at the single-cell level revealed sex differences in AD prominently in glial cells of the prefrontal cortex. In the entorhinal cortex, we observed the same genes and networks to be perturbed in opposing directions between sexes in AD relative to healthy state. Our findings contribute to growing evidence of sex differences in AD-related transcriptomic changes, which can fuel the development of therapies that may prove more effective at reversing AD pathophysiology.
Keywords: Alzheimer’s disease; RNA sequencing; Sex differences; Single-cell.
© 2021. The Author(s).
Conflict of interest statement
MS is on the advisory board of twoXAR. Other authors declare no competing financial interests.
Figures




References
-
- Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Primers. 2015;1(1):1–18. - PubMed
-
- Scheltens P, Blennow K, Breteler MMB, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer’s disease. Lancet. 2016;388(10043):505–517. - PubMed
-
- Prince MJ, Comas-Herrera A, Knapp M, Guerchet MM, Karagiannidou M. World Alzheimer report 2016—improving healthcare for people living with dementia: coverage, quality and costs now and in the future. 11/6/2020; Available from: https://kclpure.kcl.ac.uk/portal/en/publications/world-alzheimer-report-...
-
- Bureau UC. An Aging World: 2015 [Internet]. The United States Census Bureau. [cited 2021 Mar 15]. 11/6/2020 Available from: https://www.census.gov/library/publications/2016/demo/P95-16-1.html
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

