Lysis-Hi-C as a method to study polymicrobial communities and eDNA
- PMID: 34669257
- PMCID: PMC9215119
- DOI: 10.1111/1755-0998.13535
Lysis-Hi-C as a method to study polymicrobial communities and eDNA
Abstract
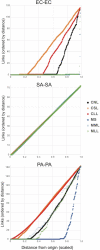
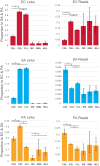
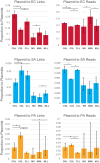
Microbes interact in natural communities in a spatially structured manner, particularly in biofilms and polymicrobial infections. While next generation sequencing approaches provide powerful insights into diversity, metabolic capacity, and mutational profiles of these communities, they generally fail to recover in situ spatial proximity between distinct genotypes in the interactome. Hi-C is a promising method that has assisted in analysing complex microbiomes, by creating chromatin cross-links in cells, that aid in identifying adjacent DNA, to improve de novo assembly. This study explored a modified Hi-C approach involving an initial lysis phase prior to DNA cross-linking, to test whether adjacent cell chromatin can be cross-linked, anticipating that this could provide a new avenue for study of spatial-mutational dynamics in structured microbial communities. An artificial polymicrobial mixture of Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli was lysed for 1-18 h, then prepared for Hi-C. A murine biofilm infection model was treated with sonication, mechanical lysis, or chemical lysis before Hi-C. Bioinformatic analyses of resulting Hi-C interspecies chromatin links showed that while microbial species differed from one another, generally lysis significantly increased links between species and increased the distance of Hi-C links within species, while also increasing novel plasmid-chromosome links. The success of this modified lysis-Hi-C protocol in creating extracellular DNA links is a promising first step toward a new lysis-Hi-C based method to recover genotypic microgeography in polymicrobial communities, with potential future applications in diseases with localized resistance, such as cystic fibrosis lung infections and chronic diabetic ulcers.
Keywords: DNA cross-linking; Hi-C; biofilm structure; genomics; polymicrobial infections.
© 2021 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


Similar articles
-
Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection.mBio. 2017 Jul 18;8(4):e00873-17. doi: 10.1128/mBio.00873-17. mBio. 2017. PMID: 28720732 Free PMC article.
-
A Pseudomonas aeruginosa Antimicrobial Affects the Biogeography but Not Fitness of Staphylococcus aureus during Coculture.mBio. 2021 Mar 30;12(2):e00047-21. doi: 10.1128/mBio.00047-21. mBio. 2021. PMID: 33785630 Free PMC article.
-
Molecular Determinants of the Thickened Matrix in a Dual-Species Pseudomonas aeruginosa and Enterococcus faecalis Biofilm.Appl Environ Microbiol. 2017 Oct 17;83(21):e01182-17. doi: 10.1128/AEM.01182-17. Print 2017 Nov 1. Appl Environ Microbiol. 2017. PMID: 28842537 Free PMC article.
-
Release mechanisms and molecular interactions of Pseudomonas aeruginosa extracellular DNA.Appl Microbiol Biotechnol. 2020 Aug;104(15):6549-6564. doi: 10.1007/s00253-020-10687-9. Epub 2020 Jun 4. Appl Microbiol Biotechnol. 2020. PMID: 32500267 Review.
-
Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: insights through network and database construction.Crit Rev Microbiol. 2019 Sep-Nov;45(5-6):712-728. doi: 10.1080/1040841X.2019.1700209. Epub 2019 Dec 13. Crit Rev Microbiol. 2019. PMID: 31835971 Review.
References
-
- Barnes, H. E. , Liu, G. , Weston, C. Q. , King, P. , Pham, L. K. , Waltz, S. , Helzer, K. T. , Day, L. , Sphar, D. , Yamamoto, R. T. , & Forsyth, R. A. (2014). Selective microbial genomic DNA isolation using restriction endonucleases. PLoS One, 9(10), e109061. 10.1371/journal.pone.0109061 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

