Use of Molecular Methods To Detect Shigella and Infer Phenotypic Resistance in a Shigella Treatment Study
- PMID: 34669456
- PMCID: PMC8769730
- DOI: 10.1128/JCM.01774-21
Use of Molecular Methods To Detect Shigella and Infer Phenotypic Resistance in a Shigella Treatment Study
Abstract
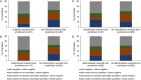
Molecular diagnostic methods improve the detection of Shigella, yet their ability to detect Shigella drug resistance on direct stool specimens is less clear. We tested 673 stool specimens from a Shigella treatment study in Bangladesh, including 154 culture-positive stool specimens and their paired Shigella isolates. We utilized a TaqMan array card that included quantitative PCR (qPCR) assays for 24 enteropathogens and 36 antimicrobial resistance (AMR) genes. Shigella was detected by culture in 23% of stool specimens (154/673), while qPCR detected Shigella at diarrhea-associated quantities in 49% (329/673; P < 0.05). qPCR for AMR genes on the Shigella isolates yielded >94% sensitivity and specificity compared with the phenotypic susceptibility results for azithromycin and ampicillin. The performance for trimethoprim-sulfamethoxazole susceptibility was less robust, and the assessment of ciprofloxacin was limited because most isolates were resistant. The detection of AMR genes in direct stool specimens generally yielded low specificities for predicting the resistance of the paired isolate, whereas the sensitivity and negative predictive values for predicting susceptibility were often higher. For example, the detection of ermB or mphA in stool yielded a specificity of 56% but a sensitivity of 91% and a negative predictive value of 91% versus the paired isolate's azithromycin resistance result. Patients who received azithromycin prior to presentation were universally culture negative (0/112); however, qPCR still detected Shigella at diarrhea-associated quantities in 34/112 (30%). In sum, molecular diagnostics on direct stool specimens greatly increase the diagnostic yield for Shigella, including in the setting of prior antibiotics. The molecular detection of drug resistance genes in direct stool specimens had low specificity for confirming resistance but could potentially "rule out" macrolide resistance.
Keywords: Shigella; molecular methods; phenotypic resistance.
Figures


References
-
- World Health Organization. 2005. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. World Health Organization, Geneva, Switzerland.
-
- Rahman M, Shoma S, Rashid H, El Arifeen S, Baqui AH, Siddique AK, Nair GB, Sack DA. 2007. Increasing spectrum in antimicrobial resistance of Shigella isolates in Bangladesh: resistance to azithromycin and ceftriaxone and decreased susceptibility to ciprofloxacin. J Health Popul Nutr 25:158–167. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

