Disrupted circadian oscillations in type 2 diabetes are linked to altered rhythmic mitochondrial metabolism in skeletal muscle
- PMID: 34669477
- PMCID: PMC8528429
- DOI: 10.1126/sciadv.abi9654
Disrupted circadian oscillations in type 2 diabetes are linked to altered rhythmic mitochondrial metabolism in skeletal muscle
Abstract
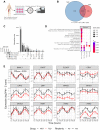
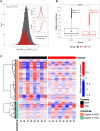
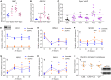
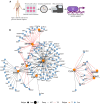
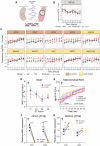
Circadian rhythms are generated by an autoregulatory feedback loop of transcriptional activators and repressors. Circadian rhythm disruption contributes to type 2 diabetes (T2D) pathogenesis. We elucidated whether altered circadian rhythmicity of clock genes is associated with metabolic dysfunction in T2D. Transcriptional cycling of core-clock genes BMAL1, CLOCK, and PER3 was altered in skeletal muscle from individuals with T2D, and this was coupled with reduced number and amplitude of cycling genes and disturbed circadian oxygen consumption. Inner mitochondria–associated genes were enriched for rhythmic peaks in normal glucose tolerance, but not T2D, and positively correlated with insulin sensitivity. Chromatin immunoprecipitation sequencing identified CLOCK and BMAL1 binding to inner-mitochondrial genes associated with insulin sensitivity, implicating regulation by the core clock. Inner-mitochondria disruption altered core-clock gene expression and free-radical production, phenomena that were restored by resveratrol treatment. We identify bidirectional communication between mitochondrial function and rhythmic gene expression, processes that are disturbed in diabetes.
Figures



References
-
- Mokhlesi B., Temple K. A., Tjaden A. H., Edelstein S. L., Utzschneider K. M., Nadeau K. J., Hannon T. S., Sam S., Barengolts E., Manchanda S., Ehrmann D. A., Van Cauter E.; Rise Consortium , Association of self-reported sleep and circadian measures with glycemia in adults with prediabetes or recently diagnosed untreated type 2 diabetes. Diabetes Care 42, 1326–1332 (2019). - PMC - PubMed
-
- Shan Z., Li Y., Zong G., Guo Y., Li J., Manson J. E., Hu F. B., Willett W. C., Schernhammer E. S., Bhupathiraju S. N., Rotating night shift work and adherence to unhealthy lifestyle in predicting risk of type 2 diabetes: Results from two large US cohorts of female nurses. BMJ 363, k4641 (2018). - PMC - PubMed
-
- Tan X., Chapman C. D., Cedernaes J., Benedict C., Association between long sleep duration and increased risk of obesity and type 2 diabetes: A review of possible mechanisms. Sleep Med. Rev. 40, 127–134 (2018). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

