RefPlantNLR is a comprehensive collection of experimentally validated plant disease resistance proteins from the NLR family
- PMID: 34669691
- PMCID: PMC8559963
- DOI: 10.1371/journal.pbio.3001124
RefPlantNLR is a comprehensive collection of experimentally validated plant disease resistance proteins from the NLR family
Abstract
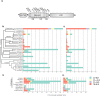
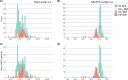
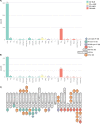
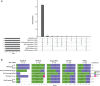
Reference datasets are critical in computational biology. They help define canonical biological features and are essential for benchmarking studies. Here, we describe a comprehensive reference dataset of experimentally validated plant nucleotide-binding leucine-rich repeat (NLR) immune receptors. RefPlantNLR consists of 481 NLRs from 31 genera belonging to 11 orders of flowering plants. This reference dataset has several applications. We used RefPlantNLR to determine the canonical features of functionally validated plant NLRs and to benchmark 5 NLR annotation tools. This revealed that although NLR annotation tools tend to retrieve the majority of NLRs, they frequently produce domain architectures that are inconsistent with the RefPlantNLR annotation. Guided by this analysis, we developed a new pipeline, NLRtracker, which extracts and annotates NLRs from protein or transcript files based on the core features found in the RefPlantNLR dataset. The RefPlantNLR dataset should also prove useful for guiding comparative analyses of NLRs across the wide spectrum of plant diversity and identifying understudied taxa. We hope that the RefPlantNLR resource will contribute to moving the field beyond a uniform view of NLR structure and function.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: The authors receive funding from industry on NLR biology.
Figures

References
-
- de Araújo AC, Fonseca FCDA, Cotta MG, Alves GSC, Miller RNG. Plant NLR receptor proteins and their potential in the development of durable genetic resistance to biotic stresses. Biotechnol Res Innov. 2020. [cited 2020 Jun 15]. doi: 10.1016/j.biori.2020.01.002 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

