UbiBrowser 2.0: a comprehensive resource for proteome-wide known and predicted ubiquitin ligase/deubiquitinase-substrate interactions in eukaryotic species
- PMID: 34669962
- PMCID: PMC8728189
- DOI: 10.1093/nar/gkab962
UbiBrowser 2.0: a comprehensive resource for proteome-wide known and predicted ubiquitin ligase/deubiquitinase-substrate interactions in eukaryotic species
Abstract
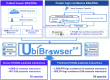
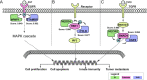
As an important post-translational modification, ubiquitination mediates ∼80% of protein degradation in eukaryotes. The degree of protein ubiquitination is tightly determined by the delicate balance between specific ubiquitin ligase (E3)-mediated ubiquitination and deubiquitinase-mediated deubiquitination. In 2017, we developed UbiBrowser 1.0, which is an integrated database for predicted human proteome-wide E3-substrate interactions. Here, to meet the urgent requirement of proteome-wide E3/deubiquitinase-substrate interactions (ESIs/DSIs) in multiple organisms, we updated UbiBrowser to version 2.0 (http://ubibrowser.ncpsb.org.cn). Using an improved protocol, we collected 4068/967 known ESIs/DSIs by manual curation, and we predicted about 2.2 million highly confident ESIs/DSIs in 39 organisms, with >210-fold increase in total data volume. In addition, we made several new features in the updated version: (i) it allows exploring proteins' upstream E3 ligases and deubiquitinases simultaneously; (ii) it has significantly increased species coverage; (iii) it presents a uniform confidence scoring system to rank predicted ESIs/DSIs. To facilitate the usage of UbiBrowser 2.0, we also redesigned the web interface for exploring these known and predicted ESIs/DSIs, and added functions of 'Browse', 'Download' and 'Application Programming Interface'. We believe that UbiBrowser 2.0, as a discovery tool, will contribute to the study of protein ubiquitination and the development of drug targets for complex diseases.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

