Hybrid immunity improves B cells and antibodies against SARS-CoV-2 variants
- PMID: 34670266
- PMCID: PMC8674140
- DOI: 10.1038/s41586-021-04117-7
Hybrid immunity improves B cells and antibodies against SARS-CoV-2 variants
Abstract
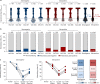
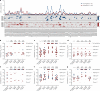
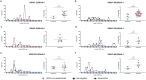
The emergence of SARS-CoV-2 variants is jeopardizing the effectiveness of current vaccines and limiting the application of monoclonal antibody-based therapy for COVID-19 (refs. 1,2). Here we analysed the memory B cells of five naive and five convalescent people vaccinated with the BNT162b2 mRNA vaccine to investigate the nature of the B cell and antibody response at the single-cell level. Almost 6,000 cells were sorted, over 3,000 cells produced monoclonal antibodies against the spike protein and more than 400 cells neutralized the original SARS-CoV-2 virus first identified in Wuhan, China. The B.1.351 (Beta) and B.1.1.248 (Gamma) variants escaped almost 70% of these antibodies, while a much smaller portion was impacted by the B.1.1.7 (Alpha) and B.1.617.2 (Delta) variants. The overall loss of neutralization was always significantly higher in the antibodies from naive people. In part, this was due to the IGHV2-5;IGHJ4-1 germline, which was found only in people who were convalescent and generated potent and broadly neutralizing antibodies. Our data suggest that people who are seropositive following infection or primary vaccination will produce antibodies with increased potency and breadth and will be able to better control emerging SARS-CoV-2 variants.
© 2021. The Author(s).
Conflict of interest statement
R.R. is an employee of the GSK group of companies. E.A., I.P., N.M., P.P., E.P., C.D.S., C.S. and R.R. are listed as inventors of full-length human mAbs described in Italian patent applications no. 102020000015754 filed on 30 June 2020, 102020000018955 filed on 3 August 2020 and 102020000029969 filed on 4 December 2020, and the international patent system number PCT/IB2021/055755 filed on 28 June 2021. All patents were submitted by Fondazione Toscana Life Sciences, Siena, Italy. The remaining authors declare no competing interests.
Figures



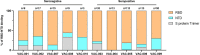

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

