Apoptosis reprogramming triggered by splicing inhibitors sensitizes multiple myeloma cells to Venetoclax treatment
- PMID: 34670358
- PMCID: PMC9152954
- DOI: 10.3324/haematol.2021.279276
Apoptosis reprogramming triggered by splicing inhibitors sensitizes multiple myeloma cells to Venetoclax treatment
Abstract
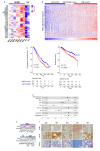
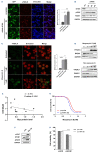
Identification of novel vulnerabilities in the context of therapeutic resistance is emerging as a key challenge for cancer treatment. Recent studies have detected pervasive aberrant splicing in cancer cells, supporting its targeting for novel therapeutic strategies. Here, we evaluated the expression of several spliceosome machinery components in multiple myeloma (MM) cells and the impact of splicing modulation on tumor cell growth and viability. A comprehensive gene expression analysis confirmed the reported deregulation of spliceosome machinery components in MM cells, compared to normal plasma cells from healthy donors, with its pharmacological and genetic modulation resulting in impaired growth and survival of MM cell lines and patient-derived malignant plasma cells. Consistent with this, transcriptomic analysis revealed deregulation of BCL2 family members, including decrease of anti-apoptotic long form of myeloid cell leukemia-1 (MCL1) expression, as crucial for "priming" MM cells for Venetoclax activity in vitro and in vivo, irrespective of t(11;14) status. Overall, our data provide a rationale for supporting the clinical use of splicing modulators as a strategy to reprogram apoptotic dependencies and make all MM patients more vulnerable to BCL2 inhibitors.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

