The complete mitochondrial genome of Ophiocordyceps gracilis and its comparison with related species
- PMID: 34670626
- PMCID: PMC8527695
- DOI: 10.1186/s43008-021-00081-z
The complete mitochondrial genome of Ophiocordyceps gracilis and its comparison with related species
Abstract
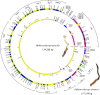
In this study, the complete mitochondrial genome of O. gracilis was sequenced and assembled before being compared with related species. As the second largest mitogenome reported in the family Ophiocordycipitaceae, the mitogenome of O. gracilis (voucher OG201301) is a circular DNA molecule of 134,288 bp that contains numerous introns and longer intergenomic regions. UCA was detected as anticodon in tRNA-Sec of O. gracilis, while comparative mitogenome analysis of nine Ophiocordycipitaceae fungi indicated that the order and contents of PCGs and rRNA genes were considerably conserved and could descend from a common ancestor in Ophiocordycipitaceae. In addition, the expansion of mitochondrial organization, introns, gene length, and order of O. gracilis were determined to be similar to those of O. sinensis, which indicated common mechanisms underlying adaptive evolution in O. gracilis and O. sinensis. Based on the mitochondrial gene dataset (15 PCGs and 2 RNA genes), a close genetic relationship between O. gracilis and O. sinensis was revealed through phylogenetic analysis. This study is the first to investigate the molecular evolution, phylogenetic pattern, and genetic structure characteristics of mitogenome in O. gracilis. Based on the obtained results, the mitogenome of O. gracilis can increase understanding of the genetic diversity and evolution of cordycipitoid fungi.
Keywords: Mitochondrial genome; Ophiocordyceps gracilis; Ophiocordycipitaceae; Phylogenetic analysis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Borstler B, Raab PA, Thiery O, Morton JB, Redecker D. Genetic diversity of the arbuscular mycorrhizal fungus Glomus intraradices as determined by mitochondrial large subunit rRNA gene sequences is considerably higher than previously expected. New Phytol. 2008;180(2):452–465. doi: 10.1111/j.1469-8137.2008.02574.x. - DOI - PubMed
Grants and funding
- 2020KY01008/Middle-aged and Young Teachers' Basic Ability Promotion Project of Guangxi
- XJ2020G048/Graduate Research and Innovation Projects of Xinjiang Province
- 31860077/National Natural Science Foundation of China
- 2016D01C039/Natural Science Foundation of Xinjiang Province
- 2017M613017/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Research Materials
