Multidrug-Resistant Methicillin-Resistant Staphylococcus aureus Associated with Bacteremia and Monocyte Evasion, Rio de Janeiro, Brazil
- PMID: 34670645
- PMCID: PMC8544994
- DOI: 10.3201/eid2711.210097
Multidrug-Resistant Methicillin-Resistant Staphylococcus aureus Associated with Bacteremia and Monocyte Evasion, Rio de Janeiro, Brazil
Abstract
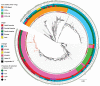
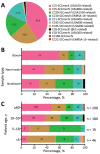
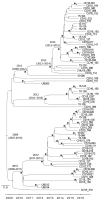
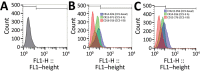
We typed 600 methicillin-resistant Staphylococcus aureus (MRSA) isolates collected in 51 hospitals in the Rio de Janeiro, Brazil, metropolitan area during 2014-2017. We found that multiple new clonal complex (CC) 5 sequence types had replaced previously dominant MRSA lineages in hospitals. Whole-genome analysis of 208 isolates revealed an emerging sublineage of multidrug-resistant MRSA, sequence type 105, staphylococcal cassette chromosome mec II, spa t002, which we designated the Rio de Janeiro (RdJ) clone. Using molecular clock analysis, we hypothesized that this lineage began to expand in the Rio de Janeiro metropolitan area in 2009. Multivariate analysis supported an association between bloodstream infections and the CC5 lineage that includes the RdJ clone. Compared with other closely related isolates, representative isolates of the RdJ clone more effectively evaded immune function related to monocytic cells, as evidenced by decreased phagocytosis rate and increased numbers of viable unphagocytosed (free) bacteria after in vitro exposure to monocytes.
Keywords: Brazil; MRSA; MRSA and other staphylococci; Rio de Janeiro; Staphylococcus aureus; antimicrobial resistance; bacteremia; bacteria; bacterial infection; bloodstream infections; drug resistance; methicillin-resistant Staphylococcus aureus; molecular epidemiology; monocyte evasion; monocytes; multidrug-resistance; phagocytosis; phylogenetics.
Figures


References
-
- Silva-Carvalho MC, Bonelli RR, Souza RR, Moreira S, dos Santos LCG, de Souza Conceição M, et al. Emergence of multiresistant variants of the community-acquired methicillin-resistant Staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2009;65:300–5. 10.1016/j.diagmicrobio.2009.07.023 - DOI - PubMed
-
- Chamon RC, Ribeiro SD, da Costa TM, Nouér SA, Dos Santos KRN. Complete substitution of the Brazilian endemic clone by other methicillin-resistant Staphylococcus aureus lineages in two public hospitals in Rio de Janeiro, Brazil. Braz J Infect Dis. 2017;21:185–9. 10.1016/j.bjid.2016.09.015 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

