NERO: a biomedical named-entity (recognition) ontology with a large, annotated corpus reveals meaningful associations through text embedding
- PMID: 34671039
- PMCID: PMC8528865
- DOI: 10.1038/s41540-021-00200-x
NERO: a biomedical named-entity (recognition) ontology with a large, annotated corpus reveals meaningful associations through text embedding
Abstract
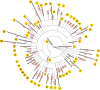
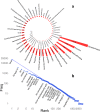
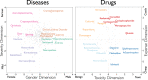
Machine reading (MR) is essential for unlocking valuable knowledge contained in millions of existing biomedical documents. Over the last two decades1,2, the most dramatic advances in MR have followed in the wake of critical corpus development3. Large, well-annotated corpora have been associated with punctuated advances in MR methodology and automated knowledge extraction systems in the same way that ImageNet4 was fundamental for developing machine vision techniques. This study contributes six components to an advanced, named entity analysis tool for biomedicine: (a) a new, Named Entity Recognition Ontology (NERO) developed specifically for describing textual entities in biomedical texts, which accounts for diverse levels of ambiguity, bridging the scientific sublanguages of molecular biology, genetics, biochemistry, and medicine; (b) detailed guidelines for human experts annotating hundreds of named entity classes; (c) pictographs for all named entities, to simplify the burden of annotation for curators; (d) an original, annotated corpus comprising 35,865 sentences, which encapsulate 190,679 named entities and 43,438 events connecting two or more entities; (e) validated, off-the-shelf, named entity recognition (NER) automated extraction, and; (f) embedding models that demonstrate the promise of biomedical associations embedded within this corpus.
© 2021. The Author(s).
Conflict of interest statement
B.B.K. is a founder of Dock Therapeutics, Inc. The remaining authors declare no competing interests.
Figures

References
-
- Banko, M. & Brill, E. Scaling to very very large corpora for natural language disambiguation. In Proc. 39th Annual Meeting on Association for Computational Linguistics 26–33 (Association for Computational Linguistics, 2001).
-
- Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intell. Syst. 2009;24:8–12. doi: 10.1109/MIS.2009.36. - DOI
-
- Deng, J. et al. ImageNet: a large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, 2009).
-
- Wijffels, J. & Okazaki, N. crfsuite: Conditional Random Fields for Labelling Sequential Data in Natural Language Processing based on CRFsuite: a fast implementation of Conditional Random Fields (CRFs) https://github.com/bnosac/crfsuite (2007–2018).
Publication types
Grants and funding
- R01 HL122712/HL/NHLBI NIH HHS/United States
- BB/D00425X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BEP17028/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U01 HL108634/HL/NHLBI NIH HHS/United States
- K12 HL143959/HL/NHLBI NIH HHS/United States
- BB/G000662/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E018025/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F008228/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D006503/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P50 MH094267/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources

