Late Quaternary dynamics of Arctic biota from ancient environmental genomics
- PMID: 34671161
- PMCID: PMC8636272
- DOI: 10.1038/s41586-021-04016-x
Late Quaternary dynamics of Arctic biota from ancient environmental genomics
Erratum in
-
Author Correction: Late Quaternary dynamics of Arctic biota from ancient environmental genomics.Nature. 2022 Mar;603(7903):E31. doi: 10.1038/s41586-022-04628-x. Nature. 2022. PMID: 35296866 Free PMC article. No abstract available.
-
Author Correction: Late Quaternary dynamics of Arctic biota from ancient environmental genomics.Nature. 2022 Oct;610(7931):E5. doi: 10.1038/s41586-022-05359-9. Nature. 2022. PMID: 36163291 Free PMC article. No abstract available.
Abstract
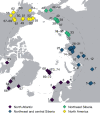
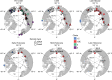
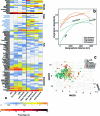
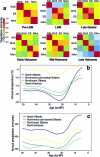
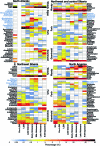
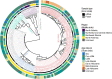
During the last glacial-interglacial cycle, Arctic biotas experienced substantial climatic changes, yet the nature, extent and rate of their responses are not fully understood1-8. Here we report a large-scale environmental DNA metagenomic study of ancient plant and mammal communities, analysing 535 permafrost and lake sediment samples from across the Arctic spanning the past 50,000 years. Furthermore, we present 1,541 contemporary plant genome assemblies that were generated as reference sequences. Our study provides several insights into the long-term dynamics of the Arctic biota at the circumpolar and regional scales. Our key findings include: (1) a relatively homogeneous steppe-tundra flora dominated the Arctic during the Last Glacial Maximum, followed by regional divergence of vegetation during the Holocene epoch; (2) certain grazing animals consistently co-occurred in space and time; (3) humans appear to have been a minor factor in driving animal distributions; (4) higher effective precipitation, as well as an increase in the proportion of wetland plants, show negative effects on animal diversity; (5) the persistence of the steppe-tundra vegetation in northern Siberia enabled the late survival of several now-extinct megafauna species, including the woolly mammoth until 3.9 ± 0.2 thousand years ago (ka) and the woolly rhinoceros until 9.8 ± 0.2 ka; and (6) phylogenetic analysis of mammoth environmental DNA reveals a previously unsampled mitochondrial lineage. Our findings highlight the power of ancient environmental metagenomics analyses to advance understanding of population histories and long-term ecological dynamics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
When did mammoths go extinct?Nature. 2022 Dec;612(7938):E1-E3. doi: 10.1038/s41586-022-05416-3. Epub 2022 Nov 30. Nature. 2022. PMID: 36450914 Free PMC article. No abstract available.
Similar articles
-
Pre-extinction Demographic Stability and Genomic Signatures of Adaptation in the Woolly Rhinoceros.Curr Biol. 2020 Oct 5;30(19):3871-3879.e7. doi: 10.1016/j.cub.2020.07.046. Epub 2020 Aug 13. Curr Biol. 2020. PMID: 32795436
-
Potential plant extinctions with the loss of the Pleistocene mammoth steppe.Nat Commun. 2025 Jan 14;16(1):645. doi: 10.1038/s41467-024-55542-x. Nat Commun. 2025. PMID: 39809751 Free PMC article.
-
Species-specific responses of Late Quaternary megafauna to climate and humans.Nature. 2011 Nov 2;479(7373):359-64. doi: 10.1038/nature10574. Nature. 2011. PMID: 22048313 Free PMC article.
-
Ancient and modern environmental DNA.Philos Trans R Soc Lond B Biol Sci. 2015 Jan 19;370(1660):20130383. doi: 10.1098/rstb.2013.0383. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25487334 Free PMC article. Review.
-
Past changes in Arctic terrestrial ecosystems, climate and UV radiation.Ambio. 2004 Nov;33(7):398-403. doi: 10.1579/0044-7447-33.7.398. Ambio. 2004. PMID: 15573568 Review.
Cited by
-
The MetaInvert soil invertebrate genome resource provides insights into below-ground biodiversity and evolution.Commun Biol. 2023 Dec 8;6(1):1241. doi: 10.1038/s42003-023-05621-4. Commun Biol. 2023. PMID: 38066075 Free PMC article.
-
A 2-million-year-old ecosystem in Greenland uncovered by environmental DNA.Nature. 2022 Dec;612(7939):283-291. doi: 10.1038/s41586-022-05453-y. Epub 2022 Dec 7. Nature. 2022. PMID: 36477129 Free PMC article.
-
Holocene shifts in marine mammal distributions around Northern Greenland revealed by sedimentary ancient DNA.Nat Commun. 2025 May 15;16(1):4543. doi: 10.1038/s41467-025-59731-0. Nat Commun. 2025. PMID: 40374632 Free PMC article.
-
Ancient DNA Reveals Maternal Philopatry of the Northeast Eurasian Brown Bear (Ursus arctos) Population during the Holocene.Genes (Basel). 2022 Oct 27;13(11):1961. doi: 10.3390/genes13111961. Genes (Basel). 2022. PMID: 36360198 Free PMC article.
-
soibean: High-Resolution Taxonomic Identification of Ancient Environmental DNA Using Mitochondrial Pangenome Graphs.Mol Biol Evol. 2024 Oct 4;41(10):msae203. doi: 10.1093/molbev/msae203. Mol Biol Evol. 2024. PMID: 39361595 Free PMC article.
References
-
- Binney H, et al. Vegetation of Eurasia from the last glacial maximum to present: key biogeographic patterns. Quat. Sci. Rev. 2017;157:80–97. doi: 10.1016/j.quascirev.2016.11.022. - DOI
-
- Bigelow, N. H. Climate change and Arctic ecosystems: 1. Vegetation changes north of 55°N between the last glacial maximum, mid-Holocene, and present. J. Geophys. Res. 108, 10.1029/2002jd002558 (2003).
-
- Stuart AJ. Late Quaternary megafaunal extinctions on the continents: a short review. Geol. J. 2015;50:338–363. doi: 10.1002/gj.2633. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

