Genomic Changes and Genetic Divergence of Vibrio alginolyticus Under Phage Infection Stress Revealed by Whole-Genome Sequencing and Resequencing
- PMID: 34671325
- PMCID: PMC8521149
- DOI: 10.3389/fmicb.2021.710262
Genomic Changes and Genetic Divergence of Vibrio alginolyticus Under Phage Infection Stress Revealed by Whole-Genome Sequencing and Resequencing
Abstract
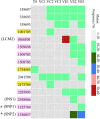
Bacteriophages (phages) and their bacterial hosts were the most abundant and genetically highly diverse organisms on the earth. In this study, a series of phage-resistant mutant (PRM) strains derived from Vibrio alginolyticus were isolated and Infrequent-restriction-site PCR (IRS-PCR) was used to investigate the genetic diversity of the PRM strains. Phenotypic variations of eight PRM strains were analyzed using profiles of utilizing carbon sources and chemical sensitivity. Genetic variations of eight PRM strains and coevolved V. alginolyticus populations with phages were analyzed by whole-genome sequencing and resequencing, respectively. The results indicated that eight genetically discrepant PRM stains exhibited abundant and abundant phenotypic variations. Eight PRM strains and coevolved V. alginolyticus populations (VE1, VE2, and VE3) contained numerous single nucleotide variations (SNVs) and insertions/indels (InDels) and exhibited obvious genetic divergence. Most of the SNVs and InDels in coding genes were related to the synthesis of flagellar, extracellular polysaccharide (EPS), which often served as the receptors of phage invasion. The PRM strains and the coevolved cell populations also contained frequent mutations in tRNA and rRNA genes. Two out of three coevolved populations (VE1 and VE2) contained a large mutation segment severely deconstructing gene nrdA, which was predictably responsible for the booming of mutation rate in the genome. In summary, numerous mutations and genetic divergence were detected in the genomes of V. alginolyticus PRM strains and in coevolved cell populations of V. alginolyticus under phage infection stress. The phage infection stress may provide an important force driving genomic evolution of V. alginolyticus.
Keywords: Vibrio alginolyticus; genetic divergence; genomic changes; phage-resistant mutations; phages.
Copyright © 2021 Zhou, Li, Li, Ma, Jiang, Hu, Ai and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Characterization and Genomic Analysis of ValSw3-3, a New Siphoviridae Bacteriophage Infecting Vibrio alginolyticus.J Virol. 2020 May 4;94(10):e00066-20. doi: 10.1128/JVI.00066-20. Print 2020 May 4. J Virol. 2020. PMID: 32132234 Free PMC article.
-
Closely Related Vibrio alginolyticus Strains Encode an Identical Repertoire of Caudovirales-Like Regions and Filamentous Phages.Viruses. 2020 Nov 27;12(12):1359. doi: 10.3390/v12121359. Viruses. 2020. PMID: 33261037 Free PMC article.
-
Genomic analysis and biological characterization of a novel Schitoviridae phage infecting Vibrio alginolyticus.Appl Microbiol Biotechnol. 2023 Feb;107(2-3):749-768. doi: 10.1007/s00253-022-12312-3. Epub 2022 Dec 15. Appl Microbiol Biotechnol. 2023. PMID: 36520169
-
Genomic and biological characterization of the Vibrio alginolyticus-infecting "Podoviridae" bacteriophage, vB_ValP_IME271.Virus Genes. 2019 Apr;55(2):218-226. doi: 10.1007/s11262-018-1622-8. Epub 2019 Jan 9. Virus Genes. 2019. PMID: 30627984
-
Complete genomic sequence of the Vibrio alginolyticus bacteriophage Vp670 and characterization of the lysis-related genes, cwlQ and holA.BMC Genomics. 2018 Oct 11;19(1):741. doi: 10.1186/s12864-018-5131-x. BMC Genomics. 2018. PMID: 30305030 Free PMC article.
Cited by
-
Role of Bacteriophages in the Evolution of Pathogenic Vibrios and Lessons for Phage Therapy.Adv Exp Med Biol. 2023;1404:149-173. doi: 10.1007/978-3-031-22997-8_8. Adv Exp Med Biol. 2023. PMID: 36792875 Free PMC article.
-
Evolutionary consequences of bacterial resistance to a flagellotropic phage.bioRxiv [Preprint]. 2025 May 10:2025.05.06.652435. doi: 10.1101/2025.05.06.652435. bioRxiv. 2025. PMID: 40654869 Free PMC article. Preprint.
-
Evolution of a bistable genetic system in fluctuating and nonfluctuating environments.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2322371121. doi: 10.1073/pnas.2322371121. Epub 2024 Aug 30. Proc Natl Acad Sci U S A. 2024. PMID: 39213178 Free PMC article.
References
-
- Abdelaziza M., Ibrahemab M. D., Ibrahimc M. A., Abu-Elala N. M., Abdel-Moneama D. A. (2017). Monitoring of different Vibrio species affecting marine fishes in Lake Qarun and Gulf of Suez: phenotypic and molecular characterization. Egypt. J. Aquat. Res. 43 141–146. 10.1016/j.ejar.2017.06.002 - DOI
LinkOut - more resources
Full Text Sources

