Monitoring Bacterial Conjugation by Optical Microscopy
- PMID: 34671336
- PMCID: PMC8521088
- DOI: 10.3389/fmicb.2021.750200
Monitoring Bacterial Conjugation by Optical Microscopy
Abstract
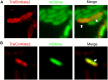
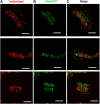
Bacterial conjugation is the main mechanism for horizontal gene transfer, conferring plasticity to the genome repertoire. This process is also the major instrument for the dissemination of antibiotic resistance genes. Hence, gathering primary information of the mechanism underlying this genetic transaction is of a capital interest. By using fluorescent protein fusions to the ATPases that power conjugation, we have been able to track the localization of these proteins in the presence and absence of recipient cells. Moreover, we have found that more than one copy of the conjugative plasmid is transferred during mating. Altogether, these findings provide new insights into the mechanism of such an important gene transfer device.
Keywords: T4SS; antibiotic resistance; bacterial conjugation; conjugative ATPases; fluorescence microscopy.
Copyright © 2021 Carranza, Menguiano, Valenzuela-Gómez, García-Cazorla, Cabezón and Arechaga.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A Streamlined Approach for Fluorescence Labelling of Low-Copy-Number Plasmids for Determination of Conjugation Frequency by Flow Cytometry.Microorganisms. 2023 Mar 29;11(4):878. doi: 10.3390/microorganisms11040878. Microorganisms. 2023. PMID: 37110299 Free PMC article.
-
Protein Transfer through an F Plasmid-Encoded Type IV Secretion System Suppresses the Mating-Induced SOS Response.mBio. 2021 Aug 31;12(4):e0162921. doi: 10.1128/mBio.01629-21. Epub 2021 Jul 13. mBio. 2021. PMID: 34253063 Free PMC article.
-
Efficient gene transfer in bacterial cell chains.mBio. 2011 Mar 15;2(2):e00027-11. doi: 10.1128/mBio.00027-11. Print 2011. mBio. 2011. PMID: 21406598 Free PMC article.
-
Towards an integrated model of bacterial conjugation.FEMS Microbiol Rev. 2015 Jan;39(1):81-95. doi: 10.1111/1574-6976.12085. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25154632 Review.
-
Bacterial conjugation: a two-step mechanism for DNA transport.Mol Microbiol. 2002 Jul;45(1):1-8. doi: 10.1046/j.1365-2958.2002.03014.x. Mol Microbiol. 2002. PMID: 12100543 Review.
Cited by
-
Chimeric systems composed of swapped Tra subunits between distantly-related F plasmids reveal striking plasticity among type IV secretion machines.bioRxiv [Preprint]. 2023 Dec 6:2023.12.05.570194. doi: 10.1101/2023.12.05.570194. bioRxiv. 2023. Update in: PLoS Genet. 2024 Mar 4;20(3):e1011088. doi: 10.1371/journal.pgen.1011088. PMID: 38106057 Free PMC article. Updated. Preprint.
-
Interference with Bacterial Conjugation and Natural Alternatives to Antibiotics: Bridging a Gap.Antibiotics (Basel). 2023 Jun 29;12(7):1127. doi: 10.3390/antibiotics12071127. Antibiotics (Basel). 2023. PMID: 37508224 Free PMC article.
-
Real-time visualisation of the intracellular dynamics of conjugative plasmid transfer.Nat Commun. 2023 Jan 18;14(1):294. doi: 10.1038/s41467-023-35978-3. Nat Commun. 2023. PMID: 36653393 Free PMC article.
-
Chimeric systems composed of swapped Tra subunits between distantly-related F plasmids reveal striking plasticity among type IV secretion machines.PLoS Genet. 2024 Mar 4;20(3):e1011088. doi: 10.1371/journal.pgen.1011088. eCollection 2024 Mar. PLoS Genet. 2024. PMID: 38437248 Free PMC article.
-
The metabolic burden associated with plasmid acquisition: An assessment of the unrecognized benefits to host cells.Bioessays. 2025 Feb;47(2):e2400164. doi: 10.1002/bies.202400164. Epub 2024 Nov 11. Bioessays. 2025. PMID: 39529437 Review.

