Identification of a Zika NS2B epitope as a biomarker for severe clinical phenotypes
- PMID: 34671736
- PMCID: PMC8459328
- DOI: 10.1039/d1md00124h
Identification of a Zika NS2B epitope as a biomarker for severe clinical phenotypes
Abstract
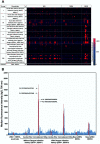
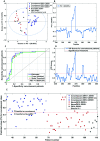
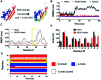
The identification of specific biomarkers for Zika infection and its clinical complications is fundamental to mitigate the infection spread, which has been associated with a broad range of neurological sequelae. We present the characterization of antibody responses in serum samples from individuals infected with Zika, presenting non-severe (classical) and severe (neurological disease) phenotypes, with high-density peptide arrays comprising the Zika NS1 and NS2B proteins. The data pinpoints one strongly IgG-targeted NS2B epitope in non-severe infections, which is absent in Zika patients, where infection progressed to the severe phenotype. This differential IgG profile between the studied groups was confirmed by multivariate data analysis. Molecular dynamics simulations and circular dichroism have shown that the peptide in solution presents itself in a sub-optimal conformation for antibody recognition, which led us to computationally engineer an artificial protein able to stabilize the NS2B epitope structure. The engineered protein was used to interrogate paired samples from mothers and their babies presenting Zika-associated microcephaly and confirmed the absence of NS2B IgG response in those samples. These findings suggest that the assessment of antibody responses to the herein identified NS2B epitope is a strong candidate biomarker for the diagnosis and prognosis of Zika-associated neurological disease.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
A patent application for the epitope has been filed by TJ, NF, FFL, TM, and ETA. A patent application for the engineered protein carrying the identified NS2B epitope has been filed by RDL, DFC and IFTV. Otherwise, the authors have declared that they have no competing interests.
Figures




References
-
- Faria N. R. Azevedo R. Kraemer M. U. G. Souza R. Cunha M. S. Hill S. C. Theze J. Bonsall M. B. Bowden T. A. Rissanen I. Rocco I. M. Nogueira J. S. Maeda A. Y. Vasami F. Macedo F. L. L. Suzuki A. Rodrigues S. G. Cruz A. C. R. Nunes B. T. Medeiros D. B. A. Rodrigues D. S. G. Queiroz A. L. N. da Silva E. V. P. Henriques D. F. da Rosa E. S. T. de Oliveira C. S. Martins L. C. Vasconcelos H. B. Casseb L. M. N. Simith D. B. Messina J. P. Abade L. Lourenco J. Alcantara L. C. J. de Lima M. M. Giovanetti M. Hay S. I. de Oliveira R. S. Lemos P. D. S. de Oliveira L. F. de Lima C. P. S. da Silva S. P. de Vasconcelos J. M. Franco L. Cardoso J. F. Vianez-Junior J. Mir D. Bello G. Delatorre E. Khan K. Creatore M. Coelho G. E. de Oliveira W. K. Tesh R. Pybus O. G. Nunes M. R. T. Vasconcelos P. F. C. Science. 2016;352:345. doi: 10.1126/science.aaf5036. - DOI - PMC - PubMed
-
- Schuler-Faccini L. Ribeiro E. M. Feitosa I. M. Horovitz D. D. Cavalcanti D. P. Pessoa A. Doriqui M. J. Neri J. I. Neto J. M. Wanderley H. Y. Cernach M. El-Husny A. S. Pone M. V. Serao C. L. Sanseverino M. T. Brazilian Medical Genetics Society-Zika Embryopathy Task F. Morb. Mortal. Wkly. Rep. 2016;65:59. doi: 10.15585/mmwr.mm6503e2. - DOI - PubMed
-
- WHO, Zika Causality Statement, https://www.who.int/emergencies/zika-virus/causality/en/, (accessed 26.03.2019, 2019)
LinkOut - more resources
Full Text Sources

