IreK-Mediated, Cell Wall-Protective Phosphorylation in Enterococcus faecalis
- PMID: 34672600
- PMCID: PMC10037947
- DOI: 10.1021/acs.jproteome.1c00635
IreK-Mediated, Cell Wall-Protective Phosphorylation in Enterococcus faecalis
Abstract
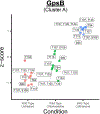
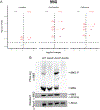
Enterococcus faecalis is a Gram-positive bacterium that is a major cause of hospital-acquired infections due, in part, to its intrinsic resistance to cell wall-active antimicrobials. One critical determinant of this resistance is the transmembrane kinase IreK, which belongs to the penicillin-binding protein and serine/threonine kinase-associated kinase family of bacterial signaling proteins involved with the regulation of cell wall homeostasis. The activity of IreK is enhanced in response to cell wall stress, but direct substrates of IreK phosphorylation, leading to antimicrobial resistance, are largely unknown. To better understand stress-modulated phosphorylation events contributing to antimicrobial resistance, wild type E. faecalis cells treated with cell wall-active antimicrobials, chlorhexidine or ceftriaxone, were examined via phosphoproteomics. Among the most prominent changes was increased phosphorylation of divisome components after both treatments, suggesting that E. faecalis modulates cell division in response to cell wall stress. Phosphorylation mediated by IreK was then determined via a similar analysis with a E. faecalis ΔireK mutant strain, revealing potential IreK substrates involved with the regulation of peptidoglycan biosynthesis and within the E. faecalis CroS/R two-component system, another signal transduction pathway that promotes antimicrobial resistance. These results reveal critical insights into the biological functions of IreK.
Keywords: Enterococcus faecalis; IreK; PASTA kinase; antimicrobial resistance; cell wall stress; mass spectrometry; phosphoproteomics.
Figures
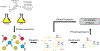

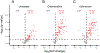

Similar articles
-
Convergence of PASTA Kinase and Two-Component Signaling in Response to Cell Wall Stress in Enterococcus faecalis.J Bacteriol. 2018 May 24;200(12):e00086-18. doi: 10.1128/JB.00086-18. Print 2018 Jun 15. J Bacteriol. 2018. PMID: 29632091 Free PMC article.
-
Phosphorylation of the cell wall hydrolase MltG in response to cell wall stress modulates resistance toward cephalosporins in Enterococcus faecalis.J Bacteriol. 2025 Aug 21;207(8):e0009925. doi: 10.1128/jb.00099-25. Epub 2025 Jul 14. J Bacteriol. 2025. PMID: 40657928 Free PMC article.
-
Reciprocal Regulation of PASTA Kinase Signaling by Differential Modification.J Bacteriol. 2019 Apr 24;201(10):e00016-19. doi: 10.1128/JB.00016-19. Print 2019 May 15. J Bacteriol. 2019. PMID: 30858297 Free PMC article.
-
The enterococcal PASTA kinase: A sentinel for cell envelope stress.Mol Oral Microbiol. 2021 Apr;36(2):132-144. doi: 10.1111/omi.12313. Epub 2020 Oct 5. Mol Oral Microbiol. 2021. PMID: 32945615 Free PMC article. Review.
-
Do Shoot the Messenger: PASTA Kinases as Virulence Determinants and Antibiotic Targets.Trends Microbiol. 2018 Jan;26(1):56-69. doi: 10.1016/j.tim.2017.06.010. Epub 2017 Jul 19. Trends Microbiol. 2018. PMID: 28734616 Free PMC article. Review.
Cited by
-
PASTA kinase signaling regulates peptidoglycan synthesis in Enterococcus faecalis by direct inhibition of UDP-N-acetylglucosamine 1-carboxyvinyl transferase activity.mBio. 2025 May 14;16(5):e0059325. doi: 10.1128/mbio.00593-25. Epub 2025 Apr 24. mBio. 2025. PMID: 40272164 Free PMC article.
-
PASTA-kinase-mediated signaling drives accumulation of the peptidoglycan synthesis protein MurAA to promote cephalosporin resistance in Enterococcus faecalis.Mol Microbiol. 2023 Dec;120(6):811-829. doi: 10.1111/mmi.15150. Epub 2023 Sep 8. Mol Microbiol. 2023. PMID: 37688380 Free PMC article.
-
1,2,3-Triazole-tethered fluoroquinolone analogues with antibacterial potential: synthesis and in vitro cytotoxicity investigations.RSC Adv. 2025 Jan 22;15(3):1896-1914. doi: 10.1039/d4ra08643k. eCollection 2025 Jan 16. RSC Adv. 2025. PMID: 39845107 Free PMC article.
-
Experimental measurement and computational prediction of bacterial Hanks-type Ser/Thr signaling system regulatory targets.Mol Microbiol. 2024 Aug;122(2):152-164. doi: 10.1111/mmi.15220. Epub 2024 Jan 3. Mol Microbiol. 2024. PMID: 38167835 Free PMC article. Review.
-
Dynamic Protein Phosphorylation in Streptococcus pyogenes during Growth, Stationary Phase, and Starvation.Microorganisms. 2024 Mar 20;12(3):621. doi: 10.3390/microorganisms12030621. Microorganisms. 2024. PMID: 38543672 Free PMC article.
References
-
- Ubeda C; Taur Y; Jenq RR; Equinda MJ; Son T; Samstein M; Viale A; Socci ND; Van Den Brink MRM; Kamboj M; Pamer EG Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest 2010, 120(12), 4332–4341. - PMC - PubMed
-
- Wells CL; Jechorek RP; Erlandsen SL Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J. Infect. Dis 1990, 162(1), 82–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

