Advances and Utility of the Human Plasma Proteome
- PMID: 34672606
- PMCID: PMC9469506
- DOI: 10.1021/acs.jproteome.1c00657
Advances and Utility of the Human Plasma Proteome
Abstract
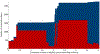
The study of proteins circulating in blood offers tremendous opportunities to diagnose, stratify, or possibly prevent diseases. With recent technological advances and the urgent need to understand the effects of COVID-19, the proteomic analysis of blood-derived serum and plasma has become even more important for studying human biology and pathophysiology. Here we provide views and perspectives about technological developments and possible clinical applications that use mass-spectrometry(MS)- or affinity-based methods. We discuss examples where plasma proteomics contributed valuable insights into SARS-CoV-2 infections, aging, and hemostasis and the opportunities offered by combining proteomics with genetic data. As a contribution to the Human Proteome Organization (HUPO) Human Plasma Proteome Project (HPPP), we present the Human Plasma PeptideAtlas build 2021-07 that comprises 4395 canonical and 1482 additional nonredundant human proteins detected in 240 MS-based experiments. In addition, we report the new Human Extracellular Vesicle PeptideAtlas 2021-06, which comprises five studies and 2757 canonical proteins detected in extracellular vesicles circulating in blood, of which 74% (2047) are in common with the plasma PeptideAtlas. Our overview summarizes the recent advances, impactful applications, and ongoing challenges for translating plasma proteomics into utility for precision medicine.
Keywords: DNA aptamers (Somascan); Human Plasma Proteome Project; Human Proteome Project; PeptideAtlas; blood; mass spectrometry; plasma; proteomics; proximity extension assays (PEA by Olink).
Conflict of interest statement
Conflict of interest
The authors declare the following competing financial interest(s): Krishnan K. Palaniappan is an employee of Freenome. Philipp E. Geyer is an employee of OmicEra Diagnostics GmbH. All other authors declare no competing financial interest.
Figures


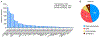
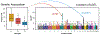
References
-
- Ignjatovic V; Geyer PE; Palaniappan KK; Chaaban JE; Omenn GS; Baker MS; Deutsch EW; Schwenk JM Mass Spectrometry-Based Plasma Proteomics: Considerations from Sample Collection to Achieving Translational Data. J. Proteome Res 2019, 18 (12), 4085–4097. 10.1021/acs.jproteome.9b00503. - DOI - PMC - PubMed
-
- Hanash S; Celis JE The Human Proteome Organization: A Mission to Advance Proteome Knowledge. Mol. Cell. Proteomics MCP 2002, 1 (6), 413–414. - PubMed
-
- Omenn GS; States DJ; Adamski M; Blackwell TW; Menon R; Hermjakob H; Apweiler R; Haab BB; Simpson RJ; Eddes JS; Kapp EA; Moritz RL; Chan DW; Rai AJ; Admon A; Aebersold R; Eng J; Hancock WS; Hefta SA; Meyer H; Paik Y-K; Yoo J-S; Ping P; Pounds J; Adkins J; Qian X; Wang R; Wasinger V; Wu CY; Zhao X; Zeng R; Archakov A; Tsugita A; Beer I; Pandey A; Pisano M; Andrews P; Tammen H; Speicher DW; Hanash SM Overview of the HUPO Plasma Proteome Project: Results from the Pilot Phase with 35 Collaborating Laboratories and Multiple Analytical Groups, Generating a Core Dataset of 3020 Proteins and a Publicly-Available Database. Proteomics 2005, 5 (13), 3226–3245. 10.1002/pmic.200500358. - DOI - PubMed
-
- Legrain P; Aebersold R; Archakov A; Bairoch A; Bala K; Beretta L; Bergeron J; Borchers CH; Corthals GL; Costello CE; Deutsch EW; Domon B; Hancock W; He F; Hochstrasser D; Marko-Varga G; Salekdeh GH; Sechi S; Snyder M; Srivastava S; Uhlén M; Wu CH; Yamamoto T; Paik Y-K; Omenn GS The Human Proteome Project: Current State and Future Direction. Mol. Cell. Proteomics 2011, 10 (7), M111.009993. 10.1074/mcp.M111.009993. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

