Transcriptomic effects of rs4845604, an IBD and allergy-associated RORC variant, in stimulated ex vivo CD4+ T cells
- PMID: 34673799
- PMCID: PMC8530322
- DOI: 10.1371/journal.pone.0258316
Transcriptomic effects of rs4845604, an IBD and allergy-associated RORC variant, in stimulated ex vivo CD4+ T cells
Abstract
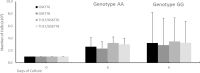
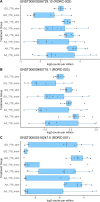
RORγt is an isoform of RORC, preferentially expressed in Th17 cells, that functions as a critical regulator of type 3 immunity. As murine Th17-driven inflammatory disease models were greatly diminished in RORC knock-out mice, this receptor was prioritised as an attractive therapeutic target for the treatment of several autoimmune diseases. Human genetic studies indicate a significant contributory role for RORC in several human disease conditions. Furthermore, genome-wide association studies (GWAS) report a significant association between inflammatory bowel disease (IBD) and the RORC regulatory variant rs4845604. To investigate if the rs4845604 variant may affect CD4+ T cell differentiation events, naïve CD4+ T cells were isolated from eighteen healthy subjects homozygous for the rs4845604 minor (A) or major (G) allele). Isolated cells from each subject were differentiated into distinct T cell lineages by culturing in either T cell maintenance medium or Th17 driving medium conditions for six days in the presence of an RORC inverse agonist (to prevent constitutive receptor activity) or an inactive diastereomer (control). Our proof of concept study indicated that genotype had no significant effect on the mean number of naïve CD4 T cells isolated, nor the frequency of Th1-like and Th17-like cells following six days of culture in any of the four culture conditions. Analysis of the derived RNA-seq count data identified genotype-driven transcriptional effects in each of the four culture conditions. Subsequent pathway enrichment analysis of these profiles reported perturbation of metabolic signalling networks, with the potential to affect the cellular detoxification response. This investigation reveals that rs4845604 genotype is associated with transcriptional effects in CD4+ T cells that may perturb immune and metabolic pathways. Most significantly, the rs4845604 GG, IBD risk associated, genotype may be associated with a differential detoxification response. This observation justifies further investigation in a larger cohort of both healthy and IBD-affected individuals.
Conflict of interest statement
We acknowledge that Glaxo SmithKline (GSK) has filed several patents in support of an RORC inverse agonist and that GSK employees may hold GSK shares. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures





References
-
- Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A. 2000;97(18):10132–7. doi: 10.1073/pnas.97.18.10132 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

