A novel NUP98-JADE2 fusion in a patient with acute myeloid leukemia resembling acute promyelocytic leukemia
- PMID: 34673934
- PMCID: PMC8791568
- DOI: 10.1182/bloodadvances.2021006064
A novel NUP98-JADE2 fusion in a patient with acute myeloid leukemia resembling acute promyelocytic leukemia
Abstract
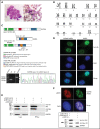
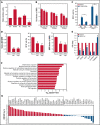
Acute promyelocytic leukemia (APL) is a specific subtype of acute myeloid leukemia (AML) characterized by block of differentiation at the promyelocytic stage and the presence of PML-RARA fusion. In rare instances, RARA is fused with other partners in variant APL. More infrequently, non-RARA genes are rearranged in AML patients resembling APL. However, the underlying disease pathogenesis in these atypical cases is largely unknown. Here, we report the identification and characterization of a NUP98- JADE2 fusion in a pediatric AML patient showing APL-like morphology and immunophenotype. Mechanistically, we showed that NUP98-JADE2 could impair all-trans retinoic acid (ATRA)-mediated transcriptional control and myeloid differentiation. Intriguingly, NUP98-JADE2 was found to alter the subcellular distribution of wild-type JADE2, whose down-regulation similarly led to attenuated ATRA-induced responses and myeloid activation, suggesting that NUP98-JADE2 may mediate JADE2 inhibition. To our knowledge, this is the first report of a NUP98-non-RAR rearrangement identified in an AML patient mimicking APL. Our findings suggest JADE2 as a novel myeloid player involved in retinoic acid-induced differentiation. Despite lacking a rearranged RARA, our findings implicate that altered retinoic acid signaling by JADE2 disruption may underlie the APL-like features in our case, corroborating the importance of this signaling in APL pathogenesis.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures
Similar articles
-
[A case of acute promyelocytic leukemia with NUP98::RARG::LINE-L2a tripartite fusion and the mechanism of resistance to all-trans retinoic acid].Zhonghua Yi Xue Za Zhi. 2025 Jul 8;105(25):2120-2123. doi: 10.3760/cma.j.cn112137-20250226-00450. Zhonghua Yi Xue Za Zhi. 2025. PMID: 40619972 Chinese.
-
Alkaloid-based regimen is beneficial for acute myeloid leukemia resembling acute promyelocytic leukemia with NUP98/RARG fusion and RUNX1 mutation: A case report.Medicine (Baltimore). 2020 Oct 2;99(40):e22488. doi: 10.1097/MD.0000000000022488. Medicine (Baltimore). 2020. PMID: 33019444 Free PMC article.
-
Recurrent RARB Translocations in Acute Promyelocytic Leukemia Lacking RARA Translocation.Cancer Res. 2018 Aug 15;78(16):4452-4458. doi: 10.1158/0008-5472.CAN-18-0840. Epub 2018 Jun 19. Cancer Res. 2018. PMID: 29921692
-
Acute myeloid leukemia with NUP98::RARG rearrangement: a case report and review of the relevant literature.Int J Hematol. 2025 Feb;121(2):265-271. doi: 10.1007/s12185-024-03881-2. Epub 2024 Dec 4. Int J Hematol. 2025. PMID: 39630349 Review.
-
Novel treatment of acute promyelocytic leukemia: As₂O₃, retinoic acid and retinoid pharmacology.Curr Pharm Biotechnol. 2013;14(9):849-58. doi: 10.2174/1389201015666140113095812. Curr Pharm Biotechnol. 2013. PMID: 24433507 Review.
Cited by
-
HBO1, a MYSTerious KAT and its links to cancer.Biochim Biophys Acta Gene Regul Mech. 2024 Sep;1867(3):195045. doi: 10.1016/j.bbagrm.2024.195045. Epub 2024 Jun 6. Biochim Biophys Acta Gene Regul Mech. 2024. PMID: 38851533 Review.
-
An Analysis of JADE2 in Non-Small Cell Lung Cancer (NSCLC).Biomedicines. 2023 Sep 19;11(9):2576. doi: 10.3390/biomedicines11092576. Biomedicines. 2023. PMID: 37761019 Free PMC article.
-
NUP98 oncofusions in myeloid malignancies: An update on molecular mechanisms and therapeutic opportunities.Hemasphere. 2024 Sep 25;8(9):e70013. doi: 10.1002/hem3.70013. eCollection 2024 Sep. Hemasphere. 2024. PMID: 39323480 Free PMC article. Review.
-
Guiding the HBO1 complex function through the JADE subunit.Nat Struct Mol Biol. 2024 Jul;31(7):1039-1049. doi: 10.1038/s41594-024-01245-2. Epub 2024 Mar 6. Nat Struct Mol Biol. 2024. PMID: 38448574 Free PMC article.
-
Novel NUP98::ASH1L Gene Fusion in Acute Myeloid Leukemia Detected by Optical Genome Mapping.Cancers (Basel). 2023 May 27;15(11):2942. doi: 10.3390/cancers15112942. Cancers (Basel). 2023. PMID: 37296904 Free PMC article.
References
-
- Wang HY, McMahon C, Ali SM, et al. . Novel FNDC3B and MECOM fusion and WT1 L378fs* 7 frameshift mutation in an acute myeloid leukaemia patient with cytomorphological and immunophenotypic features reminiscent of acute promyelocytic leukaemia. Br J Haematol. 2016;172(6):987-990. - PubMed
-
- Zhao J, Liang JW, Xue HL, et al. . The genetics and clinical characteristics of children morphologically diagnosed as acute promyelocytic leukemia. Leukemia. 2019;33(6):1387-1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

