Microbial adaptive evolution
- PMID: 34673973
- PMCID: PMC9118994
- DOI: 10.1093/jimb/kuab076
Microbial adaptive evolution
Abstract
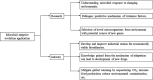
Bacterial species can adapt to significant changes in their environment by mutation followed by selection, a phenomenon known as "adaptive evolution." With the development of bioinformatics and genetic engineering, research on adaptive evolution has progressed rapidly, as have applications of the process. In this review, we summarize various mechanisms of bacterial adaptive evolution, the technologies used for studying it, and successful applications of the method in research and industry. We particularly highlight the contributions of Dr. L. O. Ingram. Microbial adaptive evolution has significant impact on our society not only from its industrial applications, but also in the evolution, emergence, and control of various pathogens.
Keywords: Adaptive evolution mechanisms; Adaptive metabolic evolution; Bacteria.
© The Author(s) 2021. Published by Oxford University Press on behalf of Society of Industrial Microbiology and Biotechnology.
Figures
References
-
- Agostini F., Sinn L., Petras D., Schipp C. J., Kubyshkin V., Berger A. A., Dorrestein P. C., Rappsilber J., Budisa N., Koksch B. (2021). Multiomics analysis provides insight into the laboratory evolution of Escherichia coli toward the metabolic usage of fluorinated indoles. ACS Central Science, 7, 81–92. 10.1021/acscentsci.0c00679 - DOI - PMC - PubMed
-
- Andersson D. I., Slechta E. S., Roth J. R. (1998). Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science, 282, 1133–1135 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources