Differential response to prey quorum signals indicates predatory specialization of myxobacteria and ability to predate Pseudomonas aeruginosa
- PMID: 34674390
- PMCID: PMC9257966
- DOI: 10.1111/1462-2920.15812
Differential response to prey quorum signals indicates predatory specialization of myxobacteria and ability to predate Pseudomonas aeruginosa
Abstract
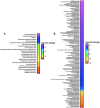
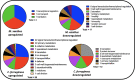
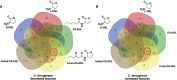
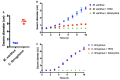
Multiomic analysis of transcriptional and metabolic responses from the predatory myxobacteria Myxococcus xanthus and Cystobacter ferrugineus exposed to prey signalling molecules of the acylhomoserine lactone and quinolone quorum signalling classes provided insight into predatory specialization. Acylhomoserine lactone quorum signals elicited a general response from both myxobacteria. We suggest that this is likely due to the generalist predator lifestyles of myxobacteria and ubiquity of acylhomoserine lactone signals. We also provide data that indicates the core homoserine lactone moiety included in all acylhomoserine lactone scaffolds to be sufficient to induce this general response. Comparing both myxobacteria, unique transcriptional and metabolic responses were observed from Cystobacter ferrugineus exposed to the quinolone signal 2-heptylquinolin-4(1H)-one (HHQ) natively produced by Pseudomonas aeruginosa. We suggest that this unique response and ability to metabolize quinolone signals contribute to the superior predation of P. aeruginosa observed from C. ferrugineus. These results further demonstrate myxobacterial eavesdropping on prey signalling molecules and provide insight into how responses to exogenous signals might correlate with prey range of myxobacteria.
© 2021 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures





References
-
- Adusumilli, R. , and Mallick, P. (2017) Data conversion with ProteoWizard msConvert. Methods Mol Biol 1550: 339–368. - PubMed
-
- Baikalov, I. , Schroder, I. , Kaczor‐Grzeskowiak, M. , Grzeskowiak, K. , Gunsalus, R.P. , and Dickerson, R.E. (1996) Structure of the Escherichia coli response regulator NarL. Biochemistry 35: 11053–11061. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

