A consensus-based ensemble approach to improve transcriptome assembly
- PMID: 34674629
- PMCID: PMC8532302
- DOI: 10.1186/s12859-021-04434-8
A consensus-based ensemble approach to improve transcriptome assembly
Abstract
Background: Systems-level analyses, such as differential gene expression analysis, co-expression analysis, and metabolic pathway reconstruction, depend on the accuracy of the transcriptome. Multiple tools exist to perform transcriptome assembly from RNAseq data. However, assembling high quality transcriptomes is still not a trivial problem. This is especially the case for non-model organisms where adequate reference genomes are often not available. Different methods produce different transcriptome models and there is no easy way to determine which are more accurate. Furthermore, having alternative-splicing events exacerbates such difficult assembly problems. While benchmarking transcriptome assemblies is critical, this is also not trivial due to the general lack of true reference transcriptomes.
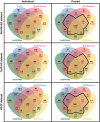
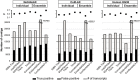
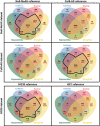
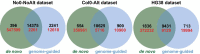
Results: In this study, we first provide a pipeline to generate a set of the simulated benchmark transcriptome and corresponding RNAseq data. Using the simulated benchmarking datasets, we compared the performance of various transcriptome assembly approaches including both de novo and genome-guided methods. The results showed that the assembly performance deteriorates significantly when alternative transcripts (isoforms) exist or for genome-guided methods when the reference is not available from the same genome. To improve the transcriptome assembly performance, leveraging the overlapping predictions between different assemblies, we present a new consensus-based ensemble transcriptome assembly approach, ConSemble.
Conclusions: Without using a reference genome, ConSemble using four de novo assemblers achieved an accuracy up to twice as high as any de novo assemblers we compared. When a reference genome is available, ConSemble using four genome-guided assemblies removed many incorrectly assembled contigs with minimal impact on correctly assembled contigs, achieving higher precision and accuracy than individual genome-guided methods. Furthermore, ConSemble using de novo assemblers matched or exceeded the best performing genome-guided assemblers even when the transcriptomes included isoforms. We thus demonstrated that the ConSemble consensus strategy both for de novo and genome-guided assemblers can improve transcriptome assembly. The RNAseq simulation pipeline, the benchmark transcriptome datasets, and the script to perform the ConSemble assembly are all freely available from: http://bioinfolab.unl.edu/emlab/consemble/ .
Keywords: Benchmarking; De novo assembly; Ensemble assembly; Genome-guided assembly; Illumina; RNAseq; Simulation; Transcriptome assembly.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Wang S, Gribskov M. Comprehensive evaluation of de novo transcriptome assembly programs and their effects on differential gene expression analysis. Bioinformatics. 2017;33(3):327–333. - PubMed
-
- Voshall A, Moriyama EN. Next-generation transcriptome assembly: strategies and performance analysis. In: Adburakhmonov IY, editor. Bioinformatics in the era of post genomics and big data. Rijeka: IntechOpen; 2018.
-
- Simonis M, Atanur SS, Linsen S, Guryev V, Ruzius FP, Game L, Lansu N, de Bruijn E, van Heesch S, Jones SJ, et al. Genetic basis of transcriptome differences between the founder strains of the rat HXB/BXH recombinant inbred panel. Genome Biol. 2012;13(4):r31. doi: 10.1186/gb-2012-13-4-r31. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

