HSP90-Specific nIR Probe Identifies Aggressive Prostate Cancers: Translation from Preclinical Models to a Human Phase I Study
- PMID: 34675120
- PMCID: PMC8742774
- DOI: 10.1158/1535-7163.MCT-21-0334
HSP90-Specific nIR Probe Identifies Aggressive Prostate Cancers: Translation from Preclinical Models to a Human Phase I Study
Abstract
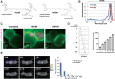
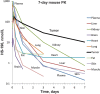
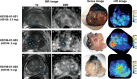
A noninvasive test to discriminate indolent prostate cancers from lethal ones would focus treatment where necessary while reducing overtreatment. We exploited the known activity of heat shock protein 90 (Hsp90) as a chaperone critical for the function of numerous oncogenic drivers, including the androgen receptor and its variants, to detect aggressive prostate cancer. We linked a near-infrared fluorescing molecule to an HSP90 binding drug and demonstrated that this probe (designated HS196) was highly sensitive and specific for detecting implanted prostate cancer cell lines with greater uptake by more aggressive subtypes. In a phase I human study, systemically administered HS196 could be detected in malignant nodules within prostatectomy specimens. Single-cell RNA sequencing identified uptake of HS196 by malignant prostate epithelium from the peripheral zone (AMACR+ERG+EPCAM+ cells), including SYP+ neuroendocrine cells that are associated with therapeutic resistance and metastatic progression. A theranostic version of this molecule is under clinical testing.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Epstein JI, Walsh PC, Carmichael M, Brendler CB. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA 1994;271:368–74. - PubMed
-
- Ploussard G, Epstein JI, Montironi R, Carroll PR, Wirth M, Grimm MO, et al. The contemporary concept of significant versus insignificant prostate cancer. Eur Urol 2011;60:291–303. - PubMed
-
- Matoso A, Epstein JI. Defining clinically significant prostate cancer on the basis of pathological findings. Histopathology 2019;74:135–45. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

