Cytokine signature and COVID-19 prediction models in the two waves of pandemics
- PMID: 34675240
- PMCID: PMC8531346
- DOI: 10.1038/s41598-021-00190-0
Cytokine signature and COVID-19 prediction models in the two waves of pandemics
Abstract
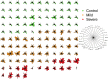
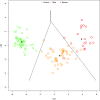
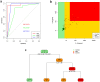
In Europe, multiple waves of infections with SARS-CoV-2 (COVID-19) have been observed. Here, we have investigated whether common patterns of cytokines could be detected in individuals with mild and severe forms of COVID-19 in two pandemic waves, and whether machine learning approach could be useful to identify the best predictors. An increasing trend of multiple cytokines was observed in patients with mild or severe/critical symptoms of COVID-19, compared with healthy volunteers. Linear Discriminant Analysis (LDA) clearly recognized the three groups based on cytokine patterns. Classification and Regression Tree (CART) further indicated that IL-6 discriminated controls and COVID-19 patients, whilst IL-8 defined disease severity. During the second wave of pandemics, a less intense cytokine storm was observed, as compared with the first. IL-6 was the most robust predictor of infection and discriminated moderate COVID-19 patients from healthy controls, regardless of epidemic peak curve. Thus, serum cytokine patterns provide biomarkers useful for COVID-19 diagnosis and prognosis. Further definition of individual cytokines may allow to envision novel therapeutic options and pave the way to set up innovative diagnostic tools.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

