β-naphthoflavone-induced upregulation of CYP1B1 expression is mediated by the preferential binding of aryl hydrocarbon receptor to unmethylated xenobiotic responsive elements
- PMID: 34676003
- PMCID: PMC8524661
- DOI: 10.3892/etm.2021.10846
β-naphthoflavone-induced upregulation of CYP1B1 expression is mediated by the preferential binding of aryl hydrocarbon receptor to unmethylated xenobiotic responsive elements
Abstract
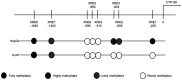
Human cytochrome P450 1 (CYP1) enzymes are transcriptionally induced by specific xenobiotics through a mechanism that involves the binding of aryl hydrocarbon receptors (AhR) to target xenobiotic responsive element (XRE) sequences. To examine the effect of DNA methylation on the AhR-mediated pathway, reverse transcription-quantitative PCR analysis was performed. β-naphthoflavone (βNF)-induced CYP1B1 expression was found to be potentiated by pre-treatment of human HepG2 liver cancer cells with 5-aza-2'-deoxycytidine, a DNA methyltransferase inhibitor, but not HuH7 cells. It was hypothesized that this increase is mediated by the demethylation of CpG sites within XRE2/XRE3 sequences, suggesting that methylation of these sequences inhibits gene expression by interfering with the binding of AhR to the target sequences. To test this hypothesis, a novel method combining the modified chromatin immunoprecipitation of AhR-XRE complexes with subsequent DNA methylation analysis of the XRE regions targeted by activated AhR was applied to both liver cancer cell lines treated with βNF. XRE2/XRE3 methylation was found to be exclusively observed in the input DNA from HepG2 cells but not in the precipitated AhR-bound DNA. Furthermore, sub-cloning and sequencing analysis revealed that the two XRE sites were unmethylated in the samples from the AhR-bound DNA even though the neighboring CpG sites were frequently methylated. To the best of our knowledge, the present study provides the first direct evidence that ligand-activated AhR preferentially binds to unmethylated XRE sequences in the context of natural chromatin. In addition, this approach can also be applied to assess the effects of DNA methylation on target sequence binding by transcription factors other than AhR.
Keywords: DNA methylation; aryl hydrocarbon receptor; cytochrome P450 family 1 subfamily B member 1; xenobiotic responsive element; β-naphthoflavone.
Copyright: © Miura et al.
Figures



References
-
- Wolf CR, Mahmood A, Henderson CJ, McLeod R, Manson MM, Neal GE, Hayes JD. Modulation of the cytochrome P450 system as a mechanism of chemoprotection. IARC Sci Publ. 1996;139:165–173. - PubMed
-
- Kress S, Greenlee WF. Cell-specific regulation of human CYP1A1 and CYP1B1 genes. Cancer Res. 1997;57:1264–1269. - PubMed
LinkOut - more resources
Full Text Sources
