Cold-chain food contamination as the possible origin of COVID-19 resurgence in Beijing
- PMID: 34676083
- PMCID: PMC7665658
- DOI: 10.1093/nsr/nwaa264
Cold-chain food contamination as the possible origin of COVID-19 resurgence in Beijing
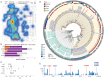
Figures
References
-
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ (29 August 2020, date last accessed).
-
- European Commission . Food Safety and the Coronavirus Disease 2019 (COVID-19). https://www.fda.gov/food/food-safety-during-emergencies/food-safety-and-... (16 August 2020, date last accessed).
LinkOut - more resources
Full Text Sources
