On the founder effect in COVID-19 outbreaks: how many infected travelers may have started them all?
- PMID: 34676089
- PMCID: PMC7543514
- DOI: 10.1093/nsr/nwaa246
On the founder effect in COVID-19 outbreaks: how many infected travelers may have started them all?
Abstract
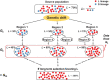
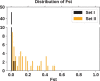
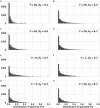
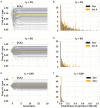
How many incoming travelers (I0 at time 0, equivalent to the 'founders' in evolutionary genetics) infected with SARS-CoV-2 who visit or return to a region could have started the epidemic of that region? I0 would be informative about the initiation and progression of epidemics. To obtain I0 , we analyze the genetic divergence among viral populations of different regions. By applying the 'individual-output' model of genetic drift to the SARS-CoV-2 diversities, we obtain I0 < 10, which could have been achieved by one infected traveler in a long-distance flight. The conclusion is robust regardless of the source population, the continuation of inputs (It for t > 0) or the fitness of the variants. With such a tiny trickle of human movement igniting many outbreaks, the crucial stage of repressing an epidemic in any region should, therefore, be the very first sign of local contagion when positive cases first become identifiable. The implications of the highly 'portable' epidemics, including their early evolution prior to any outbreak, are explored in the companion study (Ruan et al., personal communication).
Keywords: COVID-19; SARS-CoV-2; founder effect; genetic drift; population genetics.
© The Author(s) 2020. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures

References
-
- Rothman KJ, Greenland S, Lash TL. Modern Epidemiology. Philadelphia: Lippincott Williams & Wilkins, 2008.
-
- Woolhouse ME. Population biology of emerging and re-emerging pathogens. Trends Microbiol 2002; 10: S3–7. doi: 10.1016/S0966-842X(02)02428-9 - PubMed
-
- Rambaut A, Posada D, Crandall KAet al. The causes and consequences of HIV evolution. Nat Rev Genet 2004; 5: 52–61. doi: 10.1038/nrg1246 - PubMed
-
- Chen Y, Tong D, Wu CI. A new formulation of random genetic drift and its application to the evolution of cell populations. Mol Biol Evol 2017; 34: 2057–64. doi: 10.1093/molbev/msx161 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
