Cellular senescence limits translational readthrough
- PMID: 34676390
- PMCID: PMC8649927
- DOI: 10.1242/bio.058688
Cellular senescence limits translational readthrough
Abstract
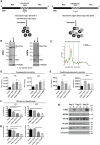
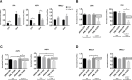
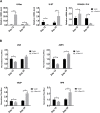
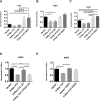
The origin and evolution of cancer cells is considered to be mainly fueled by DNA mutations. Although translation errors could also expand the cellular proteome, their role in cancer biology remains poorly understood. Tumor suppressors called caretakers block cancer initiation and progression by preventing DNA mutations and/or stimulating DNA repair. If translational errors contribute to tumorigenesis, then caretaker genes should prevent such errors in normal cells in response to oncogenic stimuli. Here, we show that the process of cellular senescence induced by oncogenes, tumor suppressors or chemotherapeutic drugs is associated with a reduction in translational readthrough (TR) measured using reporters containing termination codons withing the context of both normal translation termination or programmed TR. Senescence reduced both basal TR and TR stimulated by aminoglycosides. Mechanistically, the reduction of TR during senescence is controlled by the RB tumor suppressor pathway. Cells that escape from cellular senescence either induced by oncogenes or chemotherapy have an increased TR. Also, breast cancer cells that escape from therapy-induced senescence express high levels of AGO1x, a TR isoform of AGO1 linked to breast cancer progression. We propose that senescence and the RB pathway reduce TR limiting proteome diversity and the expression of TR proteins required for cancer cell proliferation.
Keywords: Retinoblastoma (RB) tumor suppressor; Senescence; Translation termination; Translational readthrough.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
-
- Ajore, R., Raiser, D., McConkey, M., Joud, M., Boidol, B., Mar, B., Saksena, G., Weinstock, D. M., Armstrong, S., Ellis, S. R.et al. (2017). Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 9, 498-507. 10.15252/emmm.201606660 - DOI - PMC - PubMed
-
- Baradaran-Heravi, A., Balgi, A. D., Zimmerman, C., Choi, K., Shidmoossavee, F. S., Tan, J. S., Bergeaud, C., Krause, A., Flibotte, S., Shimizu, Y.et al. (2016). Novel small molecules potentiate premature termination codon readthrough by aminoglycosides. Nucleic Acids Res. 44, 6583-6598. 10.1093/nar/gkw638 - DOI - PMC - PubMed
-
- Belin, S., Beghin, A., Solano-Gonzalez, E., Bezin, L., Brunet-Manquat, S., Textoris, J., Prats, A. C., Mertani, H. C., Dumontet, C. and Diaz, J. J. (2009). Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 4, e7147. 10.1371/journal.pone.0007147 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

