Genome-wide distribution of 5hmC in the dental pulp of mouse molars and incisors
- PMID: 34676418
- PMCID: PMC8900285
- DOI: 10.1093/jb/mvab114
Genome-wide distribution of 5hmC in the dental pulp of mouse molars and incisors
Erratum in
-
Erratum.J Biochem. 2022 Mar 31;171(4):469. doi: 10.1093/jb/mvac016. J Biochem. 2022. PMID: 35181785 Free PMC article. No abstract available.
Abstract
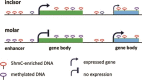
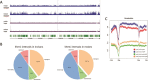
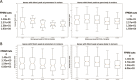
The dental pulp is critical for the production of odontoblasts to create reparative dentin. In recent years, dental pulp has become a promising source of mesenchymal stem cells that are capable of differentiating into multiple cell types. To elucidate the transcriptional control mechanisms specifying the early phases of odontoblast differentiation, we analysed the DNA demethylation pattern associated with 5-hydroxymethylcytosine (5hmC) in the primary murine dental pulp. 5hmC plays an important role in chromatin accessibility and transcriptional control by modelling a dynamic equilibrium between DNA methylation and demethylation. Our research revealed 5hmC enrichment along genes and non-coding regulatory regions associated with specific developmental pathways in the genome of mouse incisor and molar dental pulp. Although the overall distribution of 5hmC is similar, the intensity and location of the 5hmC peaks significantly differs between the incisor and molar pulp genome, indicating cell type-specific epigenetic variations. Our study suggests that the differential DNA demethylation pattern could account for the distinct regulatory mechanisms underlying the tooth-specific ontogenetic programs.
Keywords: 5-hydroxymethylcysteine (5hmC); TET enzymes; dental pulp; gene body; promoter.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Japanese Biochemical Society. All rights reserved.
Figures