RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D
- PMID: 34676613
- PMCID: PMC8740825
- DOI: 10.1002/pro.4213
RCSB Protein Data Bank: Celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D
Abstract
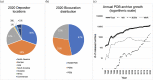
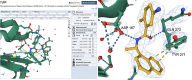
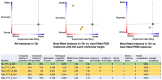
The Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), funded by the US National Science Foundation, National Institutes of Health, and Department of Energy, has served structural biologists and Protein Data Bank (PDB) data consumers worldwide since 1999. RCSB PDB, a founding member of the Worldwide Protein Data Bank (wwPDB) partnership, is the US data center for the global PDB archive housing biomolecular structure data. RCSB PDB is also responsible for the security of PDB data, as the wwPDB-designated Archive Keeper. Annually, RCSB PDB serves tens of thousands of three-dimensional (3D) macromolecular structure data depositors (using macromolecular crystallography, nuclear magnetic resonance spectroscopy, electron microscopy, and micro-electron diffraction) from all inhabited continents. RCSB PDB makes PDB data available from its research-focused RCSB.org web portal at no charge and without usage restrictions to millions of PDB data consumers working in every nation and territory worldwide. In addition, RCSB PDB operates an outreach and education PDB101.RCSB.org web portal that was used by more than 800,000 educators, students, and members of the public during calendar year 2020. This invited Tools Issue contribution describes (i) how the archive is growing and evolving as new experimental methods generate ever larger and more complex biomolecular structures; (ii) the importance of data standards and data remediation in effective management of the archive and facile integration with more than 50 external data resources; and (iii) new tools and features for 3D structure analysis and visualization made available during the past year via the RCSB.org web portal.
Keywords: Mol*; PDB; Protein Data Bank; RCSB Protein Data Bank; Worldwide Protein Data Bank; electron microscopy; macromolecular crystallography; micro-electron diffraction; open access; web-native molecular graphics.
© 2021 The Protein Society.
Figures





References
-
- Protein Data Bank . Crystallography: Protein data bank. Nat New Biol. 1971;233:223–223. - PubMed
-
- Berman HM, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Biol. 2003;10:980. - PubMed
-
- Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

