Discovery of an eIF4A Inhibitor with a Novel Mechanism of Action
- PMID: 34676755
- PMCID: PMC10103628
- DOI: 10.1021/acs.jmedchem.1c01014
Discovery of an eIF4A Inhibitor with a Novel Mechanism of Action
Abstract
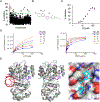
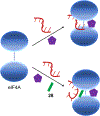
Increased protein synthesis is a requirement for malignant growth, and as a result, translation has become a pharmaceutical target for cancer. The initiation of cap-dependent translation is enzymatically driven by the eukaryotic initiation factor (eIF)4A, an ATP-powered DEAD-box RNA-helicase that unwinds the messenger RNA secondary structure upstream of the start codon, enabling translation of downstream genes. A screen for inhibitors of eIF4A ATPase activity produced an intriguing hit that, surprisingly, was not ATP-competitive. A medicinal chemistry campaign produced the novel eIF4A inhibitor 28, which decreased BJAB Burkitt lymphoma cell viability. Biochemical and cellular studies, molecular docking, and functional assays uncovered that 28 is an RNA-competitive, ATP-uncompetitive inhibitor that engages a novel pocket in the RNA groove of eIF4A and inhibits unwinding activity by interfering with proper RNA binding and suppressing ATP hydrolysis. Inhibition of eIF4A through this unique mechanism may offer new strategies for targeting this promising intersection point of many oncogenic pathways.
Conflict of interest statement
The authors declare the following competing financial interest(s): Eli Chapman is a cofounder of BioEL, Inc.
Figures


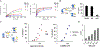

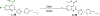

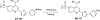
References
-
- Bhat M; Robichaud N; Hulea L; Sonenberg N; Pelletier J; Topisirovic I. Targeting the Translation Machinery in Cancer. Nat. Rev. Drug Discovery 2015, 14, 261–278. - PubMed
-
- Malka-Mahieu H; Newman M; Désaubry L; Robert C; Vagner S. Molecular Pathways: The EIF4F Translation Initiation Complex—New Opportunities for Cancer Treatment. Clin. Cancer Res 2017, 23, 21–25. - PubMed
-
- Pelletier J; Sonenberg N. The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu. Rev. Biochem 2019, 88, 307–335. - PubMed
-
- De Benedetti A; Graff JR EIF-4E Expression and Its Role in Malignancies and Metastases. Oncogene 2004, 23, 3189–3199. - PubMed
-
- Silvera D; Formenti SC; Schneider RJ Translational Control in Cancer. Nat. Rev. Cancer 2010, 10, 254–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

