Alanine-scanning mutagenesis of protein mannosyl-transferase from Streptomyces coelicolor reveals strong activity-stability correlation
- PMID: 34676818
- PMCID: PMC8698208
- DOI: 10.1099/mic.0.001103
Alanine-scanning mutagenesis of protein mannosyl-transferase from Streptomyces coelicolor reveals strong activity-stability correlation
Abstract
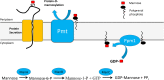
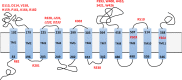
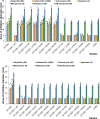
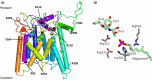
In Actinobacteria, protein O-mannosyl transferase (Pmt)-mediated protein O-glycosylation has an important role in cell envelope physiology. In S. coelicolor, defective Pmt leads to increased susceptibility to cell wall-targeting antibiotics, including vancomycin and β-lactams, and resistance to phage ϕC31. The aim of this study was to gain a deeper understanding of the structure and function of S. coelicolor Pmt. Sequence alignments and structural bioinformatics were used to identify target sites for an alanine-scanning mutagenesis study. Mutant alleles were introduced into pmt-deficient S. coelicolor strains using an integrative plasmid and scored for their ability to complement phage resistance and antibiotic hypersusceptibility phenotypes. Twenty-three highly conserved Pmt residues were each substituted for alanine. Six mutant alleles failed to complement the pmt▬ strains in either assay. Mapping the six corresponding residues onto a homology model of the three-dimensional structure of Pmt, indicated that five are positioned close to the predicted catalytic DE motif. Further mutagenesis to produce more conservative substitutions at these six residues produced Pmts that invariably failed to complement the DT1025 pmt▬ strain, indicating that strict residue conservation was necessary to preserve function. Cell fractionation and Western blotting of strains with the non-complementing pmt alleles revealed undetectable levels of the enzyme in either the membrane fractions or whole cell lysates. Meanwhile for all of the strains that complemented the antibiotic hypersusceptibility and phage resistance phenotypes, Pmt was readily detected in the membrane fraction. These data indicate a tight correlation between the activity of Pmt and its stability or ability to localize to the membrane.
Keywords: Streptomyces coelicolor; alanine-scanning mutagenesis; antibiotic susceptibility; homology model; protein mannosylation.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Streptomyces coelicolor strains lacking polyprenol phosphate mannose synthase and protein O-mannosyl transferase are hyper-susceptible to multiple antibiotics.Microbiology (Reading). 2018 Mar;164(3):369-382. doi: 10.1099/mic.0.000605. Epub 2018 Feb 1. Microbiology (Reading). 2018. PMID: 29458553 Free PMC article.
-
Characterization of the Streptomyces coelicolor Glycoproteome Reveals Glycoproteins Important for Cell Wall Biogenesis.mBio. 2019 Jun 25;10(3):e01092-19. doi: 10.1128/mBio.01092-19. mBio. 2019. PMID: 31239379 Free PMC article.
-
Lipoprotein N-acyl transferase (Lnt1) is dispensable for protein O-mannosylation by Streptomyces coelicolor.FEMS Microbiol Lett. 2014 Jan;350(1):72-82. doi: 10.1111/1574-6968.12298. Epub 2013 Oct 25. FEMS Microbiol Lett. 2014. PMID: 24117719
-
Protein O-mannosylation.Biochim Biophys Acta. 1999 Jan 6;1426(2):297-307. doi: 10.1016/s0304-4165(98)00131-7. Biochim Biophys Acta. 1999. PMID: 9878797 Review.
-
Protein-O-mannosyltransferases in virulence and development.Cell Mol Life Sci. 2008 Feb;65(4):528-44. doi: 10.1007/s00018-007-7409-z. Cell Mol Life Sci. 2008. PMID: 17975704 Free PMC article. Review.
Cited by
-
Microbial Musings - October 2021.Microbiology (Reading). 2021 Nov;167(10):001129. doi: 10.1099/mic.0.001129. Microbiology (Reading). 2021. PMID: 34846281 Free PMC article. No abstract available.
-
Structural Insights into the Protein Mannosyltransferase from Mycobacterium tuberculosis reveal a WW-Domain-Like Protein Motif in Bacteria.Commun Biol. 2025 Aug 7;8(1):1175. doi: 10.1038/s42003-025-08593-9. Commun Biol. 2025. PMID: 40775265 Free PMC article.
-
The Biological Characteristics of Mycobacterium Phage Henu3 and the Fitness Cost Associated with Its Resistant Strains.Int J Mol Sci. 2024 Aug 27;25(17):9301. doi: 10.3390/ijms25179301. Int J Mol Sci. 2024. PMID: 39273250 Free PMC article.
-
Differential substrate preferences IN ACTINOBACTERIAL protein O-MANNOSYLTRANSFERASES and alteration of protein-O-MANNOSYLATION by choice of secretion pathway.Glycobiology. 2025 Jan 13;35(1):cwae095. doi: 10.1093/glycob/cwae095. Glycobiology. 2025. PMID: 39673494 Free PMC article.
References
-
- Mahne M, Tauch A, Puhler A, Kalinowski J. The Corynebacterium glutamicum gene pmt encoding a glycosyltransferase related to eukaryotic protein-O-mannosyltransferases is essential for glycosylation of the resuscitation promoting factor (Rpf2) and other secreted proteins. FEMS Microbiol Lett. 2006;259:226–233. doi: 10.1111/j.1574-6968.2006.00269.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

