Extracellular Matrix in Synthetic Hydrogel-Based Prostate Cancer Organoids Regulate Therapeutic Response to EZH2 and DRD2 Inhibitors
- PMID: 34676924
- PMCID: PMC8820841
- DOI: 10.1002/adma.202100096
Extracellular Matrix in Synthetic Hydrogel-Based Prostate Cancer Organoids Regulate Therapeutic Response to EZH2 and DRD2 Inhibitors
Abstract
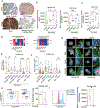
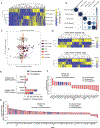
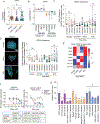
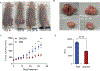
Following treatment with androgen receptor (AR) pathway inhibitors, ≈20% of prostate cancer patients progress by shedding their AR-dependence. These tumors undergo epigenetic reprogramming turning castration-resistant prostate cancer adenocarcinoma (CRPC-Adeno) into neuroendocrine prostate cancer (CRPC-NEPC). No targeted therapies are available for CRPC-NEPCs, and there are minimal organoid models to discover new therapeutic targets against these aggressive tumors. Here, using a combination of patient tumor proteomics, RNA sequencing, spatial-omics, and a synthetic hydrogel-based organoid, putative extracellular matrix (ECM) cues that regulate the phenotypic, transcriptomic, and epigenetic underpinnings of CRPC-NEPCs are defined. Short-term culture in tumor-expressed ECM differentially regulated DNA methylation and mobilized genes in CRPC-NEPCs. The ECM type distinctly regulates the response to small-molecule inhibitors of epigenetic targets and Dopamine Receptor D2 (DRD2), the latter being an understudied target in neuroendocrine tumors. In vivo patient-derived xenograft in immunocompromised mice showed strong anti-tumor response when treated with a DRD2 inhibitor. Finally, we demonstrate that therapeutic response in CRPC-NEPCs under drug-resistant ECM conditions can be overcome by first cellular reprogramming with epigenetic inhibitors, followed by DRD2 treatment. The synthetic organoids suggest the regulatory role of ECM in therapeutic response to targeted therapies in CRPC-NEPCs and enable the discovery of therapies to overcome resistance.
Keywords: chemoresistance; dopamine receptors; epigenetics; neuroendocrine; tumor microenvironment.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of Interest
V.P. and J.E.A. are employees and shareholders of Chimerix, Inc. M.J.M is an employee at Teva Pharmaceutical and this work was completed prior to his employment at Teva. L.P is an employee at Loxo Oncology at Lilly, this work was completed prior to her employment at Loxo, she is acting on her own, and these endeavors are not in any manner affiliated with Loxo Oncology at Lilly.
Figures



References
-
- Davies AH, Beltran H, Zoubeidi A, Nat. Rev. Urol 2018, 15, 271. - PubMed
-
- Siegel RL, Miller KD, Jemal A, Ca-Cancer J. Clin 2018, 68, 7. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Ca-Cancer J. Clin 2018, 68, 394. - PubMed
-
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Flechon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS, N. Engl. J. Med 2012, 367, 1187. - PubMed
-
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB Jr., Saad F, Staffurth JN, Mainwaring P, Harland S, Flaig TW, Hutson TE, Cheng T, Patterson H, Hainsworth JD, Ryan CJ, Sternberg CN, Ellard SL, Flechon A, Saleh M, Scholz M, Efstathiou E, Zivi A, Bianchini D, Loriot Y, Chieffo N, Kheoh T, Haqq CM, Scher HI, N. Engl. J. Med 2011, 364, 1995. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 5R01AI132738-04/NH/NIH HHS/United States
- 1T32EB023860-01A1/NH/NIH HHS/United States
- R01 AI132738/AI/NIAID NIH HHS/United States
- EEC-1648035/Burroughs Wellcome Fund National Science Foundation
- R37 CA241486/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- T32 EB023860/EB/NIBIB NIH HHS/United States
- DMR-1554275/National Science Foundation
- 1R01CA238745-01A1/Wellcome Leap HOPE Program National Institutes of Health
- R01 CA238745/CA/NCI NIH HHS/United States
- 5P50CA211024/NH/NIH HHS/United States
- P50 CA211024/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

