Drug target inference by mining transcriptional data using a novel graph convolutional network framework
- PMID: 34677780
- PMCID: PMC8532448
- DOI: 10.1007/s13238-021-00885-0
Drug target inference by mining transcriptional data using a novel graph convolutional network framework
Abstract
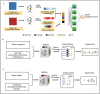
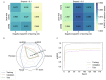
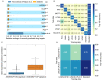
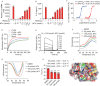
A fundamental challenge that arises in biomedicine is the need to characterize compounds in a relevant cellular context in order to reveal potential on-target or off-target effects. Recently, the fast accumulation of gene transcriptional profiling data provides us an unprecedented opportunity to explore the protein targets of chemical compounds from the perspective of cell transcriptomics and RNA biology. Here, we propose a novel Siamese spectral-based graph convolutional network (SSGCN) model for inferring the protein targets of chemical compounds from gene transcriptional profiles. Although the gene signature of a compound perturbation only provides indirect clues of the interacting targets, and the biological networks under different experiment conditions further complicate the situation, the SSGCN model was successfully trained to learn from known compound-target pairs by uncovering the hidden correlations between compound perturbation profiles and gene knockdown profiles. On a benchmark set and a large time-split validation dataset, the model achieved higher target inference accuracy as compared to previous methods such as Connectivity Map. Further experimental validations of prediction results highlight the practical usefulness of SSGCN in either inferring the interacting targets of compound, or reversely, in finding novel inhibitors of a given target of interest.
Keywords: deep learning; drug target inference; experimental verification; transcriptomics.
© 2021. The Author(s).
Figures



References
-
- Abbas AK, Trotta E, Simeonov DR, Marson A, Bluestone JA (2018) Revisiting IL-2: biology and therapeutic prospects. Sci Immunol 3:eaat1482. - PubMed
-
- André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. New Engl J Med. 2019;380:1929–1940. - PubMed
-
- Anighoro A, Bajorath J, Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. J Med Chem. 2014;57:7874–7887. - PubMed
-
- Arshad U, Pertinez H, Box H, Tatham L, Rajoli RKR, Curley P, Neary M, Sharp J, Liptrott NJ, Valentijn A, et al. Prioritization of anti-SARS-Cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin Pharmacol Ther. 2020 doi: 10.1002/cpt.1909. - DOI - PMC - PubMed
-
- Ashburn TT, Thor KB. Drug repositioning: Identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

