A Small Molecule Exploits Hidden Structural Features within the RNA Repeat Expansion That Causes c9ALS/FTD and Rescues Pathological Hallmarks
- PMID: 34677935
- PMCID: PMC9594102
- DOI: 10.1021/acschemneuro.1c00470
A Small Molecule Exploits Hidden Structural Features within the RNA Repeat Expansion That Causes c9ALS/FTD and Rescues Pathological Hallmarks
Abstract
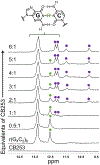
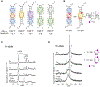
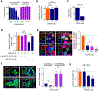
The hexanucleotide repeat expansion GGGGCC [r(G4C2)exp] within intron 1 of C9orf72 causes genetically defined amyotrophic lateral sclerosis and frontotemporal dementia, collectively named c9ALS/FTD. , the repeat expansion causes neurodegeneration via deleterious phenotypes stemming from r(G4C2)exp RNA gain- and loss-of-function mechanisms. The r(G4C2)exp RNA folds into both a hairpin structure with repeating 1 × 1 nucleotide GG internal loops and a G-quadruplex structure. Here, we report the identification of a small molecule (CB253) that selectively binds the hairpin form of r(G4C2)exp. Interestingly, the small molecule binds to a previously unobserved conformation in which the RNA forms 2 × 2 nucleotide GG internal loops, as revealed by a series of binding and structural studies. NMR and molecular dynamics simulations suggest that the r(G4C2)exp hairpin interconverts between 1 × 1 and 2 × 2 internal loops through the process of strand slippage. We provide experimental evidence that CB253 binding indeed shifts the equilibrium toward the 2 × 2 GG internal loop conformation, inhibiting mechanisms that drive c9ALS/FTD pathobiology, such as repeat-associated non-ATG translation formation of stress granules and defective nucleocytoplasmic transport in various cellular models of c9ALS/FTD.
Keywords: NMR spectroscopy; RNA; amyotrophic lateral sclerosis; bistable RNA; frontotemporal dementia; microsatellite disorders; quinazoline; repeat associate non-ATG (RAN) translation; repeat expansion; small molecules.
Figures



Similar articles
-
Conformational dynamics of RNA G4C2 and G2C4 repeat expansions causing ALS/FTD using NMR and molecular dynamics studies.Nucleic Acids Res. 2023 Jun 23;51(11):5325-5340. doi: 10.1093/nar/gkad403. Nucleic Acids Res. 2023. PMID: 37216594 Free PMC article.
-
The Hairpin Form of r(G4C2)exp in c9ALS/FTD Is Repeat-Associated Non-ATG Translated and a Target for Bioactive Small Molecules.Cell Chem Biol. 2019 Feb 21;26(2):179-190.e12. doi: 10.1016/j.chembiol.2018.10.018. Epub 2018 Nov 29. Cell Chem Biol. 2019. PMID: 30503283 Free PMC article.
-
Structural Features of Small Molecules Targeting the RNA Repeat Expansion That Causes Genetically Defined ALS/FTD.ACS Chem Biol. 2020 Dec 18;15(12):3112-3123. doi: 10.1021/acschembio.0c00049. Epub 2020 Nov 16. ACS Chem Biol. 2020. PMID: 33196168 Free PMC article.
-
All in the Family: Repeats and ALS/FTD.Trends Neurosci. 2018 May;41(5):247-250. doi: 10.1016/j.tins.2018.03.010. Trends Neurosci. 2018. PMID: 29703376 Free PMC article. Review.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
Cited by
-
Molecular basis of RNA-binding and autoregulation by the cancer-associated splicing factor RBM39.Nat Commun. 2023 Sep 4;14(1):5366. doi: 10.1038/s41467-023-40254-5. Nat Commun. 2023. PMID: 37666821 Free PMC article.
-
Conformational dynamics of RNA G4C2 and G2C4 repeat expansions causing ALS/FTD using NMR and molecular dynamics studies.Nucleic Acids Res. 2023 Jun 23;51(11):5325-5340. doi: 10.1093/nar/gkad403. Nucleic Acids Res. 2023. PMID: 37216594 Free PMC article.
-
Mapping of repeat-associated non-AUG (RAN) translation knowledge: A bibliometric analysis.Heliyon. 2024 Apr 2;10(8):e29141. doi: 10.1016/j.heliyon.2024.e29141. eCollection 2024 Apr 30. Heliyon. 2024. PMID: 38628764 Free PMC article.
-
Advances in the Structure of GGGGCC Repeat RNA Sequence and Its Interaction with Small Molecules and Protein Partners.Molecules. 2023 Aug 1;28(15):5801. doi: 10.3390/molecules28155801. Molecules. 2023. PMID: 37570771 Free PMC article. Review.
-
Therapeutic targeting of RNA for neurological and neuromuscular disease.Genes Dev. 2024 Sep 19;38(15-16):698-717. doi: 10.1101/gad.351612.124. Genes Dev. 2024. PMID: 39142832 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

