Development of a highly specific and sensitive VHH-based sandwich immunoassay for the detection of the SARS-CoV-2 nucleoprotein
- PMID: 34678315
- PMCID: PMC8526496
- DOI: 10.1016/j.jbc.2021.101290
Development of a highly specific and sensitive VHH-based sandwich immunoassay for the detection of the SARS-CoV-2 nucleoprotein
Abstract
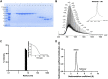
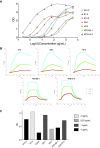
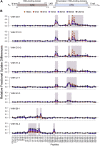
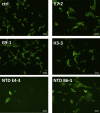
The current COVID-19 pandemic illustrates the importance of obtaining reliable methods for the rapid detection of SARS-CoV-2. A highly specific and sensitive diagnostic test able to differentiate the SARS-CoV-2 virus from common human coronaviruses is therefore needed. Coronavirus nucleoprotein (N) localizes to the cytoplasm and the nucleolus and is required for viral RNA synthesis. N is the most abundant coronavirus protein, so it is of utmost importance to develop specific antibodies for its detection. In this study, we developed a sandwich immunoassay to recognize the SARS-CoV-2 N protein. We immunized one alpaca with recombinant SARS-CoV-2 N and constructed a large single variable domain on heavy chain (VHH) antibody library. After phage display selection, seven VHHs recognizing the full N protein were identified by ELISA. These VHHs did not recognize the nucleoproteins of the four common human coronaviruses. Hydrogen Deuterium eXchange-Mass Spectrometry (HDX-MS) analysis also showed that these VHHs mainly targeted conformational epitopes in either the C-terminal or the N-terminal domains. All VHHs were able to recognize SARS-CoV-2 in infected cells or on infected hamster tissues. Moreover, the VHHs could detect the SARS variants B.1.17/alpha, B.1.351/beta, and P1/gamma. We propose that this sandwich immunoassay could be applied to specifically detect the SARS-CoV-2 N in human nasal swabs.
Keywords: COVID-19; antibody engineering; diagnostic; hydrogen-deuterium exchange; immunochemistry; nanobodies; nucleoprotein; phage display; single domain antibodies.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

