Genome-wide analyses of light-regulated genes in Aspergillus nidulans reveal a complex interplay between different photoreceptors and novel photoreceptor functions
- PMID: 34679095
- PMCID: PMC8535378
- DOI: 10.1371/journal.pgen.1009845
Genome-wide analyses of light-regulated genes in Aspergillus nidulans reveal a complex interplay between different photoreceptors and novel photoreceptor functions
Abstract
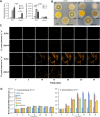
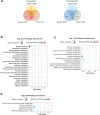
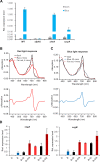
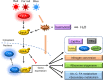
Fungi sense light of different wavelengths using blue-, green-, and red-light photoreceptors. Blue light sensing requires the "white-collar" proteins with flavin as chromophore, and red light is sensed through phytochrome. Here we analyzed genome-wide gene expression changes caused by short-term, low-light intensity illumination with blue-, red- or far-red light in Aspergillus nidulans and found that more than 1100 genes were differentially regulated. The largest number of up- and downregulated genes depended on the phytochrome FphA and the attached HOG pathway. FphA and the white-collar orthologue LreA fulfill activating but also repressing functions under all light conditions and both appear to have roles in the dark. Additionally, we found about 100 genes, which are red-light induced in the absence of phytochrome, suggesting alternative red-light sensing systems. We also found blue-light induced genes in the absence of the blue-light receptor LreA. We present evidence that cryptochrome may be part of this regulatory cue, but that phytochrome is essential for the response. In addition to in vivo data showing that FphA is involved in blue-light sensing, we performed spectroscopy of purified phytochrome and show that it responds indeed to blue light.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Red- and Blue-Light Sensing in the Plant Pathogen Alternaria alternata Depends on Phytochrome and the White-Collar Protein LreA.mBio. 2019 Apr 9;10(2):e00371-19. doi: 10.1128/mBio.00371-19. mBio. 2019. PMID: 30967462 Free PMC article.
-
Light-dependent gene activation in Aspergillus nidulans is strictly dependent on phytochrome and involves the interplay of phytochrome and white collar-regulated histone H3 acetylation.Mol Microbiol. 2015 Aug;97(4):733-45. doi: 10.1111/mmi.13062. Epub 2015 Jun 26. Mol Microbiol. 2015. PMID: 25980340
-
The Aspergillus nidulans phytochrome FphA represses sexual development in red light.Curr Biol. 2005 Oct 25;15(20):1833-8. doi: 10.1016/j.cub.2005.08.061. Curr Biol. 2005. PMID: 16243030
-
Light in the Fungal World: From Photoreception to Gene Transcription and Beyond.Annu Rev Genet. 2019 Dec 3;53:149-170. doi: 10.1146/annurev-genet-120417-031415. Epub 2019 Aug 26. Annu Rev Genet. 2019. PMID: 31451036 Review.
-
Photosensing fungi: phytochrome in the spotlight.Curr Biol. 2005 Oct 25;15(20):R829-32. doi: 10.1016/j.cub.2005.10.001. Curr Biol. 2005. PMID: 16243020 Review.
Cited by
-
Current Trends of Bacterial and Fungal Optoproteins for Novel Optical Applications.Int J Mol Sci. 2023 Sep 29;24(19):14741. doi: 10.3390/ijms241914741. Int J Mol Sci. 2023. PMID: 37834188 Free PMC article. Review.
-
Light regulation of secondary metabolism in fungi.J Biol Eng. 2023 Aug 31;17(1):57. doi: 10.1186/s13036-023-00374-4. J Biol Eng. 2023. PMID: 37653453 Free PMC article. Review.
-
White Collar 1 Modulates Oxidative Sensitivity and Virulence by Regulating the HOG1 Pathway in Fusarium asiaticum.Microbiol Spectr. 2023 Jun 15;11(3):e0520622. doi: 10.1128/spectrum.05206-22. Epub 2023 May 17. Microbiol Spectr. 2023. PMID: 37195224 Free PMC article.
-
Red and far-red light improve the antagonistic ability of Trichoderma guizhouense against phytopathogenic fungi by promoting phytochrome-dependent aerial hyphal growth.PLoS Genet. 2024 May 20;20(5):e1011282. doi: 10.1371/journal.pgen.1011282. eCollection 2024 May. PLoS Genet. 2024. PMID: 38768261 Free PMC article.
-
Structure and distribution of sensor histidine kinases in the fungal kingdom.Curr Genet. 2024 Sep 14;70(1):17. doi: 10.1007/s00294-024-01301-w. Curr Genet. 2024. PMID: 39276214
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

