The Key Differences between Human Papillomavirus-Positive and -Negative Head and Neck Cancers: Biological and Clinical Implications
- PMID: 34680354
- PMCID: PMC8533896
- DOI: 10.3390/cancers13205206
The Key Differences between Human Papillomavirus-Positive and -Negative Head and Neck Cancers: Biological and Clinical Implications
Abstract
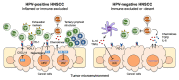
Head and neck squamous cell carcinoma (HNSCC) is a unique malignancy associated with two distinct risk factors: exposure to typical carcinogens and infection of human papillomavirus (HPV). HPV encodes the potent oncoproteins E6 and E7, which bypass many important oncogenic processes and result in cancer development. In contrast, HPV-negative HNSCC is developed through multiple mutations in diverse oncogenic driver genes. While the risk factors associated with HPV-positive and HPV-negative HNSCCs are discrete, HNSCC patients still show highly complex molecular signatures, immune infiltrations, and treatment responses even within the same anatomical subtypes. Here, we summarize the current understanding of biological mechanisms, treatment approaches, and clinical outcomes in comparison between HPV-positive and -negative HNSCCs.
Keywords: clinical outcome; clinical trials; de-escalation strategies; head and neck cancer; head and neck squamous cell carcinoma; human papillomavirus; microbiome; molecular carcinogenesis; surgery; treatment; tumor microenvironment.
Conflict of interest statement
W.C.S. consulting for Bristol Myers Squibb, Regeneron, and Merck. S.F.P. received research grant support to the institution from Merck, Bristol Myers Squib, Pfizer, Vyriad, Incyte, Actuate, Genentech, and Seattle Genetics. Steven Powell received consulting support to the institution from Bristol Myers Squibb. The other authors declare no conflict of interest.
Figures

References
-
- Pyeon D., Newton M.A., Lambert P.F., Boon J.A.D., Sengupta S., Marsit C., Woodworth C.D., Connor J.P., Haugen T., Smith E.M., et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

