Protein Unfolding: Denaturant vs. Force
- PMID: 34680512
- PMCID: PMC8533514
- DOI: 10.3390/biomedicines9101395
Protein Unfolding: Denaturant vs. Force
Abstract
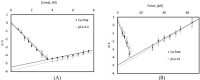
While protein refolding has been studied for over 50 years since the pioneering work of Christian Anfinsen, there have been a limited number of studies correlating results between chemical, thermal, and mechanical unfolding. The limited knowledge of the relationship between these processes makes it challenging to compare results between studies if different refolding methods were applied. Our current work compares the energetic barriers and folding rates derived from chemical, thermal, and mechanical experiments using an immunoglobulin-like domain from the muscle protein titin as a model system. This domain, I83, has high solubility and low stability relative to other Ig domains in titin, though its stability can be modulated by calcium. Our experiments demonstrated that the free energy of refolding was equivalent with all three techniques, but the refolding rates exhibited differences, with mechanical refolding having slightly faster rates. This suggests that results from equilibrium-based measurements can be compared directly but care should be given comparing refolding kinetics derived from refolding experiments that used different unfolding methods.
Keywords: chemical denaturation; immunoglobulin domain; magnetic tweezers; protein refolding; thermal denaturation; titin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Levinthal C. Are there pathways for protein folding? J. Chim. Phys. 1968;65:44–45. doi: 10.1051/jcp/1968650044. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

