Factors Regulating the Activity of LINE1 Retrotransposons
- PMID: 34680956
- PMCID: PMC8535693
- DOI: 10.3390/genes12101562
Factors Regulating the Activity of LINE1 Retrotransposons
Abstract
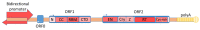
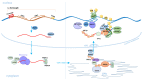
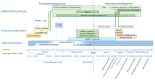
LINE-1 (L1) is a class of autonomous mobile genetic elements that form somatic mosaicisms in various tissues of the organism. The activity of L1 retrotransposons is strictly controlled by many factors in somatic and germ cells at all stages of ontogenesis. Alteration of L1 activity was noted in a number of diseases: in neuropsychiatric and autoimmune diseases, as well as in various forms of cancer. Altered activity of L1 retrotransposons for some pathologies is associated with epigenetic changes and defects in the genes involved in their repression. This review discusses the molecular genetic mechanisms of the retrotransposition and regulation of the activity of L1 elements. The contribution of various factors controlling the expression and distribution of L1 elements in the genome occurs at all stages of the retrotransposition. The regulation of L1 elements at the transcriptional, post-transcriptional and integration into the genome stages is described in detail. Finally, this review also focuses on the evolutionary aspects of L1 accumulation and their interplay with the host regulation system.
Keywords: L1 silencing; LINE-1 retrotransposons; regulation; repetitive elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

