More Than a Moggy; A Population Genetics Analysis of the United Kingdom's Non-Pedigree Cats
- PMID: 34681013
- PMCID: PMC8535647
- DOI: 10.3390/genes12101619
More Than a Moggy; A Population Genetics Analysis of the United Kingdom's Non-Pedigree Cats
Abstract
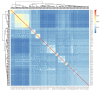
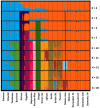
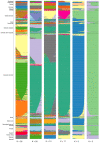
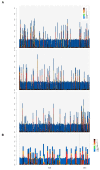
The domestic cat is one of the most popular pets in the world. It is estimated that 89-92% of domestic cats in the UK are non-pedigree Domestic shorthair (DSH), Domestic longhair (DLH), or Domestic semi-longhair cats (DSLH). Despite their popularity, little is known of the UK non-pedigree cats' population structure and breeding dynamics. Using a custom designed single nucleotide variant (SNV) array, this study investigated the population genetics of 1344 UK cats. Principal components analysis (PCA) and fastSTRUCTURE analysis verified that the UK's DSH, DLH, and DSLH cats are random-bred, rather than admixed, mix breed, or crossbred. In contrast to pedigree cats, the linkage disequilibrium of these random-bred cats was least extensive and decayed rapidly. Homozygosity by descent (HBD) analysis showed the majority of non-pedigree cats had proportionally less of their genome in HBD segments compared to pedigree cats, and that these segments were older. Together, these findings suggest that the DSH, DLH, and DSLH cats should be considered as a population of random-bred cats rather than a crossbred or pedigree-admixed cat. Unexpectedly, 19% of random-bred cat genomes displayed a higher proportion of HBD segments associated with more recent inbreeding events. Therefore, while non-pedigree cats as a whole are genetically diverse, they are not impervious to inbreeding and its health risks.
Keywords: SNV; autozygosity; cat; genotype; inbreeding; population genetics; random-bred; structure.
Conflict of interest statement
Jennifer Irving McGrath is a recipient of the Hills Pet Nutrition PhD studentship at the University of Edinburgh. Jeffrey J. Schoenebeck, Richard Mellanby, and Danielle Gunn-Moore received research support from Hills Pet Nutrition. Jeffrey A. Brockman and Regina Hollar are employees of Hills Pet Nutrition. Alison Collings is an employee of Idexx Laboratories. Roger Powell is an employee and co-founder of DragonVet Consulting Ltd. Rob D Foale and Nicola Thurley are employees of Dick White Referrals.
Figures





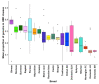
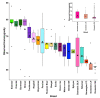
References
-
- Statista Cats in the United States Since 2000. [(accessed on 3 March 2021)]. Available online: https://www.statista.com/statistics/198102/cats-in-the-united-states-sin....
-
- PDSA PDSA Animal Welfare (PAW) Report. [(accessed on 1 February 2021)]. Available online: https://www.pdsa.org.uk/media/10540/pdsa-paw-report-2020.pdf.
-
- Lipinski M.J., Froenicke L., Baysac K.C., Billings N.C., Leutenegger C.M., Levy A.M., Longeri M., Niini T., Ozpinar H., Slater M.R., et al. The ascent of cat breeds: Genetic evaluations of breeds and worldwide random-bred populations. Genomics. 2008;91:12–21. doi: 10.1016/j.ygeno.2007.10.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- BBS/E/D/10002070/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/10002071/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/30002275/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/30002276/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

