Loss of the R2R3 MYB Transcription Factor RsMYB1 Shapes Anthocyanin Biosynthesis and Accumulation in Raphanus sativus
- PMID: 34681588
- PMCID: PMC8535906
- DOI: 10.3390/ijms222010927
Loss of the R2R3 MYB Transcription Factor RsMYB1 Shapes Anthocyanin Biosynthesis and Accumulation in Raphanus sativus
Abstract
The red or purple color of radish (Raphanus sativus L.) taproots is due to anthocyanins, which have nutritional and aesthetic value, as well as antioxidant properties. Moreover, the varied patterns and levels of anthocyanin accumulation in radish roots make them an interesting system for studying the transcriptional regulation of anthocyanin biosynthesis. The R2R3 MYB transcription factor RsMYB1 is a key positive regulator of anthocyanin biosynthesis in radish. Here, we isolated an allele of RsMYB1, named RsMYB1Short, in radish cultivars with white taproots. The RsMYB1Short allele carried a 4 bp insertion in the first exon causing a frame-shift mutation of RsMYB1, generating a truncated protein with only a partial R2 domain at the N-terminus. Unlike RsMYB1Full, RsMYB1Short was localized to the nucleus and the cytoplasm and failed to interact with their cognate partner RsTT8. Transient expression of genomic or cDNA sequences for RsMYB1Short in radish cotyledons failed to induce anthocyanin accumulation, but that for RsMYB1Full activated it. Additionally, RsMYB1Short showed the lost ability to induce pigment accumulation and to enhance the transcript level of anthocyanin biosynthetic genes, while RsMYB1Full promoted both processes when co-expressed with RsTT8 in tobacco leaves. As the result of the transient assay, co-expressing RsTT8 and RsMYB1Full, but not RsMYB1Short, also enhanced the promoter activity of RsCHS and RsDFR. We designed a molecular marker for RsMYB1 genotyping, and revealed that the RsMYB1Short allele is common in white radish cultivars, underscoring the importance of variation at the RsMYB1 locus in anthocyanin biosynthesis in the radish taproot. Together, these results indicate that the nonsense mutation of RsMYB1 generated the truncated protein, RsMYB1Short, that had the loss of ability to regulate anthocyanin biosynthesis. Our findings highlight that the frame shift mutation of RsMYB1 plays a key role in anthocyanin biosynthesis in the radish taproot.
Keywords: MBW complex; RsMYB1; anthocyanin; frameshift mutation; radish.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

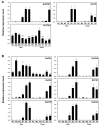



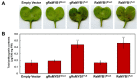





References
-
- Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

