MicroRNA-124 Alleviates Retinal Vasoregression via Regulating Microglial Polarization
- PMID: 34681723
- PMCID: PMC8538759
- DOI: 10.3390/ijms222011068
MicroRNA-124 Alleviates Retinal Vasoregression via Regulating Microglial Polarization
Abstract
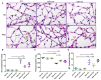
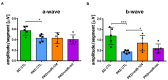
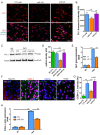
Microglial activation is implicated in retinal vasoregression of the neurodegenerative ciliopathy-associated disease rat model (i.e., the polycystic kidney disease (PKD) model). microRNA can regulate microglial activation and vascular function, but the effect of microRNA-124 (miR-124) on retinal vasoregression remains unclear. Transgenic PKD and wild-type Sprague Dawley (SD) rats received miR-124 at 8 and 10 weeks of age intravitreally. Retinal glia activation was assessed by immunofluorescent staining and in situ hybridization. Vasoregression and neuroretinal function were evaluated by quantitative retinal morphometry and electroretinography (ERG), respectively. Microglial polarization was determined by immunocytochemistry and qRT-PCR. Microglial motility was examined via transwell migration assays, wound healing assays, and single-cell tracking. Our data showed that miR-124 inhibited glial activation and improved vasoregession, as evidenced by the reduced pericyte loss and decreased acellular capillary formation. In addition, miR-124 improved neuroretinal function. miR-124 shifted microglial polarization in the PKD retina from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype by suppressing TNF-α, IL-1β, CCL2, CCL3, MHC-II, and IFN-γ and upregulating Arg1 and IL-10. miR-124 also decreased microglial motility in the migration assays. The transcriptional factor of C/EBP-α-PU.1 signaling, suppressed by miR-124 both in vivo (PKD retina) and in vitro (microglial cells), could serve as a key regulator in microglial activation and polarization. Our data illustrate that miR-124 regulates microglial activation and polarization. miR-124 inhibits pericyte loss and thereby alleviates vasoregression and ameliorates neurovascular function.
Keywords: miR-124; microglia; polarization; vasoregression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
miRNA-124 Prevents Rat Diabetic Retinopathy by Inhibiting the Microglial Inflammatory Response.Int J Mol Sci. 2023 Jan 24;24(3):2291. doi: 10.3390/ijms24032291. Int J Mol Sci. 2023. PMID: 36768614 Free PMC article.
-
Protective effect of Soluble Epoxide Hydrolase Inhibition in Retinal Vasculopathy associated with Polycystic Kidney Disease.Theranostics. 2020 Jun 22;10(17):7857-7871. doi: 10.7150/thno.43154. eCollection 2020. Theranostics. 2020. PMID: 32685025 Free PMC article.
-
MicroRNA-195 prevents hippocampal microglial/macrophage polarization towards the M1 phenotype induced by chronic brain hypoperfusion through regulating CX3CL1/CX3CR1 signaling.J Neuroinflammation. 2020 Aug 20;17(1):244. doi: 10.1186/s12974-020-01919-w. J Neuroinflammation. 2020. PMID: 32819407 Free PMC article.
-
Bu Shen Yi Sui Capsule Alleviates Neuroinflammation and Demyelination by Promoting Microglia toward M2 Polarization, Which Correlates with Changes in miR-124 and miR-155 in Experimental Autoimmune Encephalomyelitis.Oxid Med Cell Longev. 2021 Mar 16;2021:5521503. doi: 10.1155/2021/5521503. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 33815654 Free PMC article.
-
Shaping the Microglia in Retinal Degenerative Diseases Using Stem Cell Therapy: Practice and Prospects.Front Cell Dev Biol. 2021 Dec 13;9:741368. doi: 10.3389/fcell.2021.741368. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34966736 Free PMC article. Review.
Cited by
-
Neuroinflammation in retinitis pigmentosa: Therapies targeting the innate immune system.Front Immunol. 2022 Oct 27;13:1059947. doi: 10.3389/fimmu.2022.1059947. eCollection 2022. Front Immunol. 2022. PMID: 36389729 Free PMC article. Review.
-
miRNA-124 Prevents Rat Diabetic Retinopathy by Inhibiting the Microglial Inflammatory Response.Int J Mol Sci. 2023 Jan 24;24(3):2291. doi: 10.3390/ijms24032291. Int J Mol Sci. 2023. PMID: 36768614 Free PMC article.
-
Oral 8-aminoguanine against age-related retinal degeneration.Res Sq [Preprint]. 2024 May 6:rs.3.rs-4022389. doi: 10.21203/rs.3.rs-4022389/v1. Res Sq. 2024. Update in: Commun Biol. 2025 May 26;8(1):812. doi: 10.1038/s42003-025-08242-1. PMID: 38765984 Free PMC article. Updated. Preprint.
-
Neural Correlates and Molecular Mechanisms of Memory and Learning.Int J Mol Sci. 2024 Feb 27;25(5):2724. doi: 10.3390/ijms25052724. Int J Mol Sci. 2024. PMID: 38473973 Free PMC article.
-
Emerging Translational Research in Neurological and Psychiatric Diseases: From In Vitro to In Vivo Models.Int J Mol Sci. 2023 Oct 30;24(21):15739. doi: 10.3390/ijms242115739. Int J Mol Sci. 2023. PMID: 37958722 Free PMC article.
References
-
- Bellver-Landete V., Bretheau F., Mailhot B., Vallieres N., Lessard M., Janelle M.E., Vernoux N., Tremblay M.E., Fuehrmann T., Shoichet M.S., et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 2019;10:518. doi: 10.1038/s41467-019-08446-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

